Chapter 8 Major Earthquake Analysis
We apply the concepts in the previous chapters to the series of annual counts of major earthquakes (i.e. magnitude 7 and above) from the textbook.
The data can be read into R
eq_dat <- read.table("http://www.hmms-for-time-series.de/second/data/earthquakes.txt")We use the following packages for the analysis
# Load packages
library(kableExtra)
library(tidyverse)
library(gridExtra)
library(momentuHMM)
library(rstan)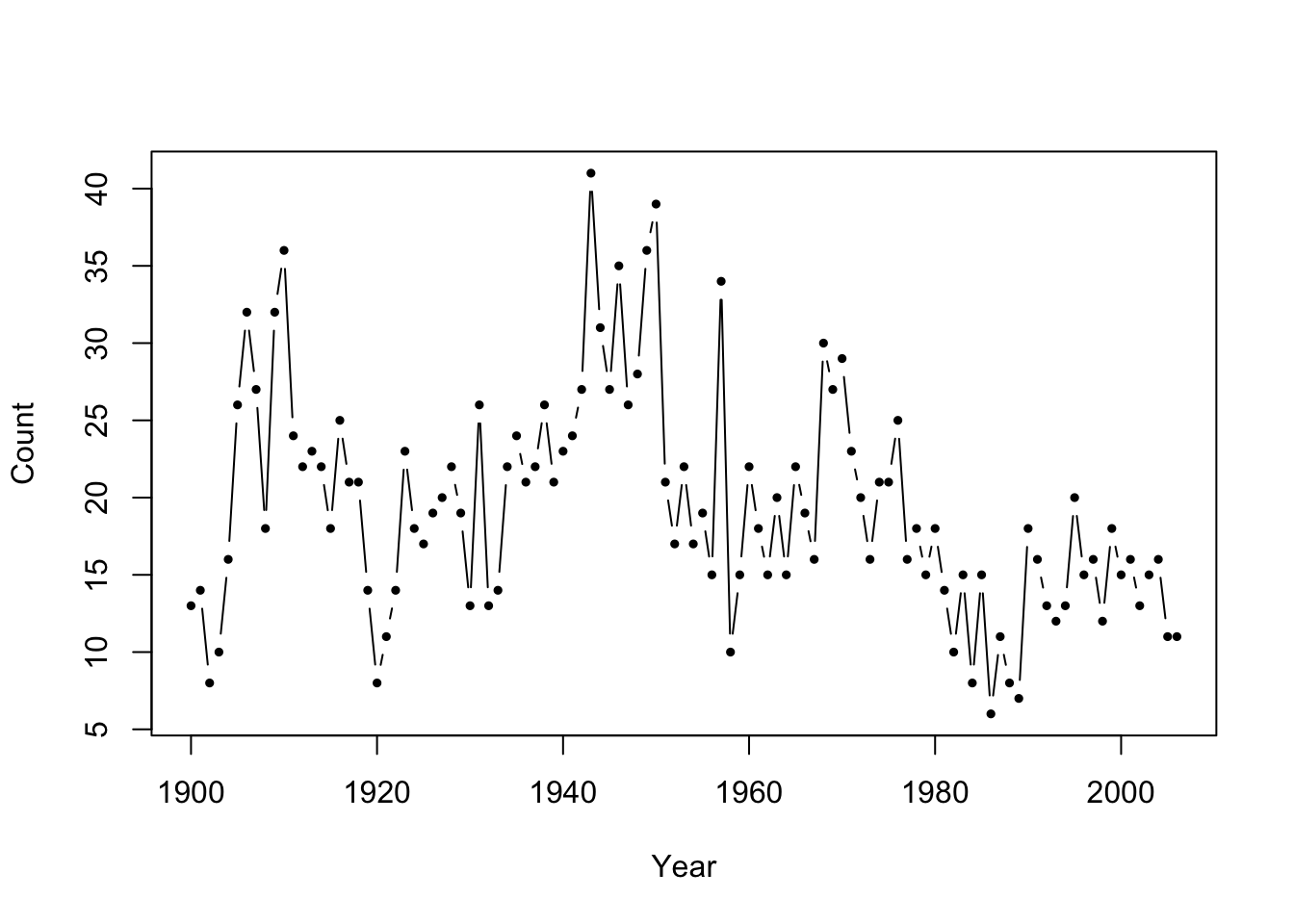
Figure 8.1: Number of major earthquakes worldwide from 1900-2006.
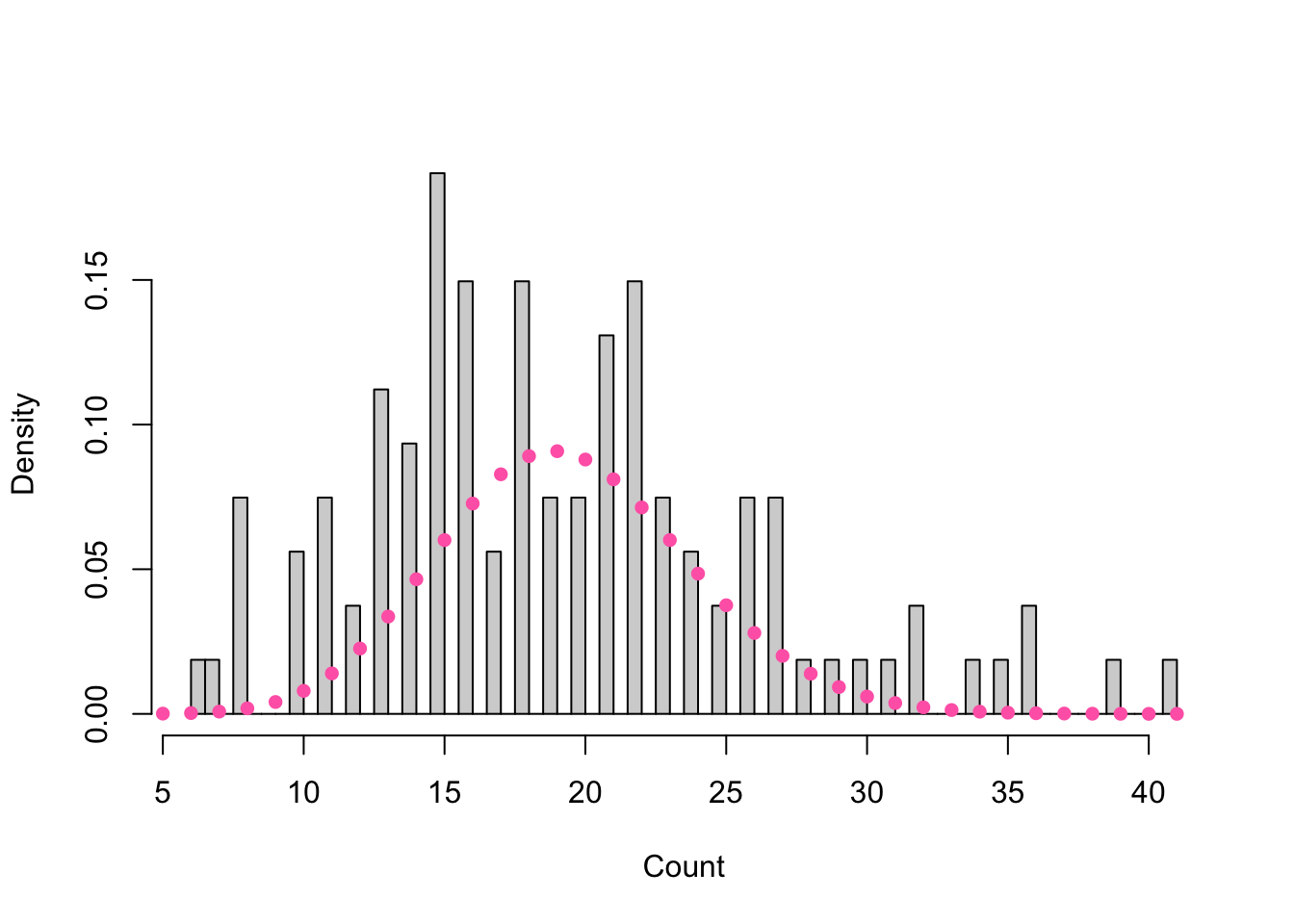
Figure 8.2: Histogram of major earthquakes overlaid with a Poisson density of mean equal to the observed counts.
| mean | variance |
|---|---|
| 19.36 | 51.57 |
Fitting a Poisson Mixture Distribution
Note: See Preliminaries
We may consider fitting a Poisson mixture distribution since earthquake counts are unbounded and there appears to be overdispersion as evident by the multimodal peaks in Figure 8.2 and sample variance \(s^2 \approx 52 > 19 \approx \bar{x}\) in Table 8.1.
Suppose \(m=3\) and the three components are Poisson-distributed with means \(\lambda_1, \lambda_2\), and \(\lambda_3\). Let \(\delta_1\), \(\delta_2\), and \(\delta_3\) be the respective mixing parameters. The mixture distribution \(p\) is given by
\[p(x) = \delta_1 \frac{\lambda_1^x e^{-\lambda_1}}{x!} + \delta_2 \frac{\lambda_2^x e^{-\lambda_2}}{x!} + \delta_3 \frac{\lambda_3^x e^{-\lambda_3}}{x!}\]
The likelihood is given by
\[L(\lambda_1, \lambda_2, \lambda_3, \delta_1, \delta_2|x_1, \dots, x_n) = \prod_{i=1}^n \left( \delta_1 \frac{\lambda_1^{x_i} e^{-\lambda_1}}{x_i!} + \delta_2 \frac{\lambda_2^{x_i} e^{-\lambda_2}}{x_i!} + (1 - \delta_1 - \delta_2) \frac{\lambda_3^{x_i} e^{-\lambda_3}}{x_i!} \right)\]
The log-likelihood can be maximized using the unconstrained optimizer nlm on reparameterized parameters by the following:
Step 1: Reparameterize the “natural parameters” \(\boldsymbol{\delta}\) and \(\boldsymbol{\lambda}\) into “working parameters” \(\eta_i = \log \lambda_i \qquad{(i = 1, \dots, m)}\) and \(\tau_i = \log \left(\frac{\delta_i}{1 - \sum_{j=2}^m \delta_j}\right) (i = 2, \dots, m)\)
n2w <- function(lambda, delta)log(c(lambda, delta[-1]/(1-sum(delta[-1]))))Step 2: Compute the negative log-likelihood using the working parameters
mllk <- function(wpar, x){
zzz <- w2n(wpar)
-sum(log(outer(x, zzz$lambda, dpois)%*%zzz$delta))}Step 3: Transform the parameters back into natural parameters
w2n <- function(wpar){
m <- (length(wpar)+1)/2
lambda <- exp(wpar[1:m])
delta <- exp(c(0, wpar[(m+1):(2*m-1)]))
return(list(lambda=lambda, delta=delta/sum(delta)))}Hence, for a Poisson mixture distribution with means \(\lambda_1, = 10, \lambda_2 = 20, \lambda_3 = 25\) and probabilities \(\delta_1 = \delta_2 = \delta_3 = \frac{1}{3}\),
wpar <- n2w(c(10, 20, 25), c(1,1,1)/3)w2n(nlm(mllk, wpar, eq_dat$Count)$estimate)we obtain the following parameter estimates
| lambda | delta |
|---|---|
| 12.73573 | 0.2775329 |
| 19.78515 | 0.5928037 |
| 31.62940 | 0.1296634 |
and the negative log likelihood
nlm(mllk, wpar, eq_dat$Count)$minimum## [1] 356.8489Note: See Table 1.2 and Figure 1.4 of the textbook for a comparison of fitted mixture models with varying number of components.
Fitting a Poisson-HMM by Numerical Maximization
Note: See Introduction to Hidden Markov Models and Appendix A of the textbook.
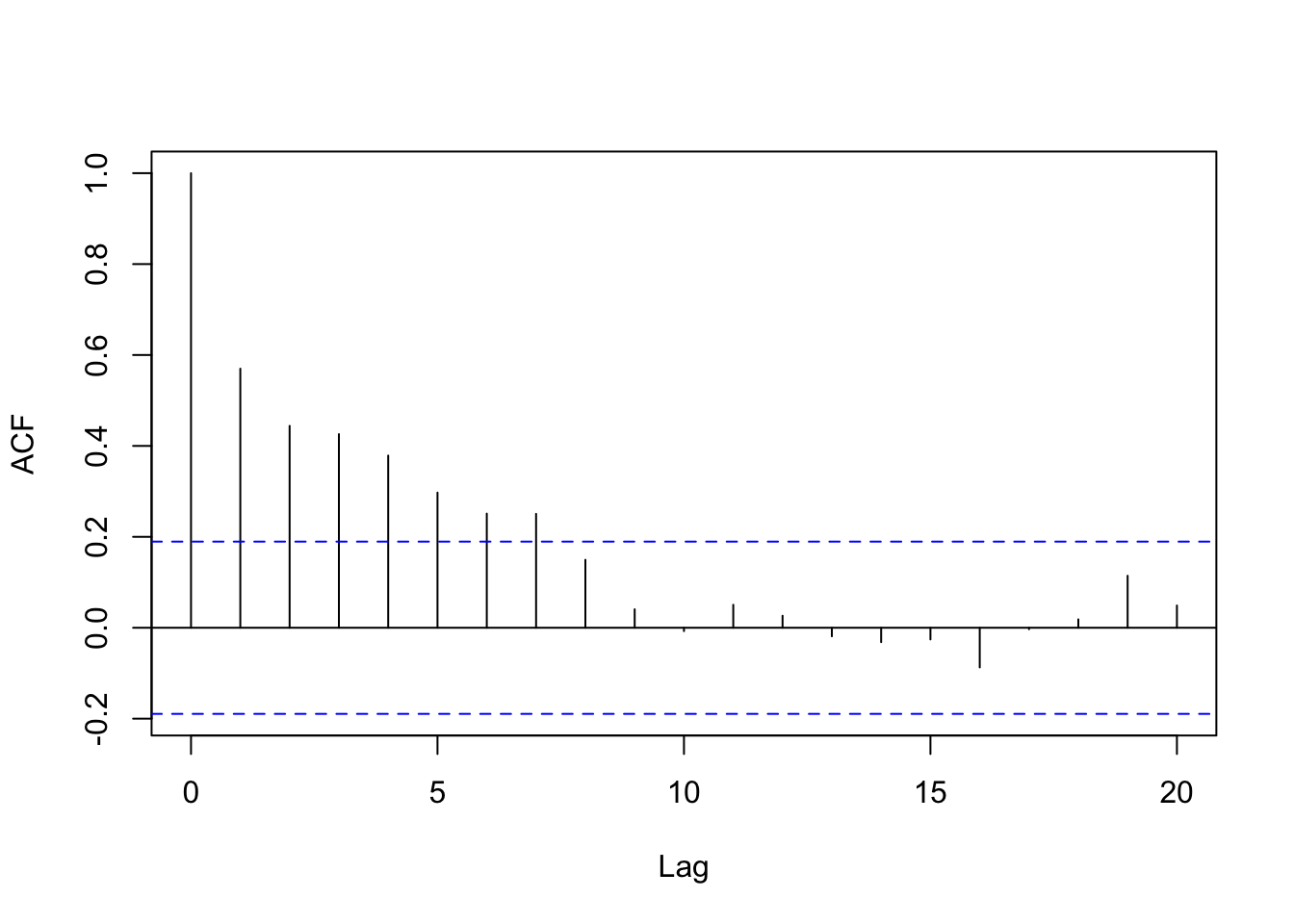
Figure 8.3: Autocorrelation plot for major earthquakes.
Instead of fitting an (independent) Poisson mixture distribution, we may consider fitting a Poisson-HMM to account for the strong, positive serial dependence as evident in Figure 8.3.
Step 1: Reparameterize the “natural parameters” \(\boldsymbol{\Gamma}, \boldsymbol{\lambda}\), and \(\boldsymbol{\delta}\) to the “working parameters”.
Set \[\eta_i = \log \lambda_i \qquad{(i = 1, \dots, m)}\]
\[\nu_{ij} = \begin{cases} 1 & \qquad{\text{for } i = j} \\ \log(\gamma_{ij}) & \qquad{\text{for } i \neq j} \end{cases}\]
\[\delta_i = \begin{cases} \log \left(\frac{(\delta_2, \dots, \delta_m)}{\delta_1}\right) & \qquad{\text{if not stationary}}\\ \text{NULL} & \qquad{\text{otherwise}} \end{cases}\]
See Reparameterization to Avoid Constraints.
pois.HMM.pn2pw <- function(m,lambda,gamma,delta=NULL,stationary=TRUE){
# eta = log(lambda)
tlambda <- log(lambda)
if(m==1) return(tlambda)
# For i = j, nu_ii = 1
foo <- log(gamma/diag(gamma))
# For i != j, nu_ij = log(gamma_ij)
tgamma <- as.vector(foo[!diag(m)])
# If stationary, set to pi = null
if(stationary) {tdelta <- NULL}
# Otherwise, pi = log((delta_2, ..., delta_m)/delta_1)
else {tdelta <- log(delta[-1]/delta[1])}
parvect <- c(tlambda,tgamma,tdelta)
return(parvect)
}Note: pois.HMM.pn2pw takes in arguments m number of states (numeric), lambda Poisson means (vector), delta initial probabilities, and stationary (TRUE/FALSE). By default, the MC is assumed to be stationary with the initial distribution equal to the stationary distribution. The function returns a vector containing the working parameters.
Step 2: Compute the log-likelihood by the forward algorithm using the scaling strategy.
The general algorithm is:
\(w_1 \leftarrow \boldsymbol{\delta P} (x_1) \boldsymbol{1'};\) \(\boldsymbol{\phi}_1 \leftarrow \boldsymbol{\delta P} (x_1);\) \(l \leftarrow \log w_1\)
for \(t=2, 3, \dots, T\)
\[\begin{align} \boldsymbol{v} &\leftarrow \boldsymbol{\phi}_{t-1} \boldsymbol{\Gamma P} (x_t)\\ u & \leftarrow \boldsymbol{v 1'}\\ l &\leftarrow l + \log u\\ \boldsymbol{\phi}_t & \leftarrow \frac{\boldsymbol{v}}{u} \end{align}\]
See Scaling the Likelihood Computation and Note (4).
pois.HMM.lforward <- function(x,mod){
n <- length(x)
lalpha <- matrix(NA,mod$m,n)
# At time t=1
foo <- mod$delta*dpois(x[1],mod$lambda)
sumfoo <- sum(foo)
lscale <- log(sumfoo)
foo <- foo/sumfoo
lalpha[,1] <- lscale+log(foo)
# For t > 1
for (i in 2:n){
foo <- foo%*%mod$gamma*dpois(x[i],mod$lambda)
sumfoo <- sum(foo)
lscale <- lscale+log(sumfoo)
foo <- foo/sumfoo
lalpha[,i] <- log(foo)+lscale
}
return(lalpha)
}Note: pois.HMM.lforward takes in arguments x data and mod list consisting of m states, lambda means for the Poisson state-dependent distribution, and delta initial probabilities.
Step 3: Maximize the log-likelihood/Minimize the negative log-likelihood
The function nlm in R performs the minimization of any function using a Newton-type algorithm. In order to minimize the negative log-likelihood, we first write a function that compute the negative log-likelihood from the working parameters. The same forward algorithm scheme applies.
pois.HMM.mllk <- function(parvect,x,m,stationary=TRUE,...)
{
if(m==1) return(-sum(dpois(x,exp(parvect),log=TRUE)))
n <- length(x)
pn <- pois.HMM.pw2pn(m,parvect,stationary=stationary)
foo <- pn$delta*dpois(x[1],pn$lambda)
sumfoo <- sum(foo)
lscale <- log(sumfoo)
foo <- foo/sumfoo
for (i in 2:n)
{
if(!is.na(x[i])){P<-dpois(x[i],pn$lambda)}
else {P<-rep(1,m)}
foo <- foo %*% pn$gamma*P
sumfoo <- sum(foo)
lscale <- lscale+log(sumfoo)
foo <- foo/sumfoo
}
mllk <- -lscale
return(mllk)
}Step 4: Transform the parameters back into natural parameters.
Set
\[\hat{\lambda_i} = \exp(\hat{\eta_i}) \qquad{(i = 1, \dots, m)}\]
\[\hat{\gamma}_{ij} = \frac{\exp(\hat{\nu_{ik}})}{\sum_{k=1, k \neq i}^m\exp(\hat{\nu_ik})} \qquad{\text{for } (i, j = 1, \dots, m)}\]
\[\hat{\boldsymbol{\delta}} = \begin{cases} \boldsymbol{1} (\boldsymbol{I} - \hat{\boldsymbol{\Gamma}} + \boldsymbol{U})^{-1}& \qquad{\text{if stationary}}\\ \frac{(1, \exp(\hat{\delta_2}), \dots, \exp(\hat{\delta}_m))}{1 + \exp(\hat{\delta_2}) + \dots + \exp(\hat{\delta}_m)} & \qquad{\text{otherwise}} \end{cases}\]
pois.HMM.pw2pn <- function(m,parvect,stationary=TRUE){
lambda <- exp(parvect[1:m])
gamma <- diag(m)
if (m==1) return(list(lambda=lambda,gamma=gamma,delta=1))
gamma[!gamma] <- exp(parvect[(m+1):(m*m)])
gamma <- gamma/apply(gamma,1,sum)
if(stationary){delta<-solve(t(diag(m)-gamma+1),rep(1,m))}
else {foo<-c(1,exp(parvect[(m*m+1):(m*m+m-1)]))
delta<-foo/sum(foo)}
return(list(lambda=lambda,gamma=gamma,delta=delta))
}Note: pois.HMM.pw2pn takes in arguments m number of states, parvect working parameters (vector), and stationary (TRUE/FALSE). By default, the MC is assumed to be stationary with the initial distribution equal to the stationary distribution. The function returns a list containing the natural parameters.
We can combine the parts above into the pois.HMM.mle function.
pois.HMM.mle <- function(x,m,lambda0,gamma0,delta0=NULL,stationary=TRUE,...){
parvect0 <- pois.HMM.pn2pw(m,lambda0,gamma0,delta0,stationary=stationary)
mod <- nlm(pois.HMM.mllk,parvect0,x=x,m=m,stationary=stationary)
pn <- pois.HMM.pw2pn(m=m,mod$estimate,stationary=stationary)
mllk <- mod$minimum
np <- length(parvect0)
AIC <- 2*(mllk+np)
n <- sum(!is.na(x))
BIC <- 2*mllk+np*log(n)
list(m=m,lambda=pn$lambda,gamma=pn$gamma,delta=pn$delta,code=mod$code,mllk=mllk,AIC=AIC,BIC=BIC)
}Hence, for a three-state Poisson-HMM with the following parameters
\[\boldsymbol{\lambda} = (10, 20, 25)\]
\[\boldsymbol{\Gamma} = \begin{pmatrix} 0.8 & 0.1 & 0.1\\ 0.1 & 0.8 & 0.1\\ 0.1 & 0.1 & 0.8 \end{pmatrix}\]
m = 3
lambda = c(10, 20, 25)
gamma = matrix(c(0.8,0.1,0.1,0.1,0.8,0.1,0.1,0.1,0.8),3,3,byrow=TRUE)if we assume that the MC is stationary
eq_init_s <- list(m=m,
lambda=lambda,
gamma = gamma)
(mod3s<-pois.HMM.mle(eq_dat$Count, eq_init_s$m,eq_init_s$lambda,eq_init_s$gamma,stationary=TRUE))## $m
## [1] 3
##
## $lambda
## [1] 13.14573 19.72101 29.71437
##
## $gamma
## [,1] [,2] [,3]
## [1,] 9.546243e-01 0.0244426 0.02093313
## [2,] 4.976679e-02 0.8993673 0.05086592
## [3,] 1.504495e-09 0.1966420 0.80335799
##
## $delta
## [1] 0.4436420 0.4044983 0.1518597
##
## $code
## [1] 1
##
## $mllk
## [1] 329.4603
##
## $AIC
## [1] 676.9206
##
## $BIC
## [1] 700.976and if we do not assume stationary and that \(\boldsymbol{\delta} = (\frac{1}{3}, \frac{1}{3}, \frac{1}{3})\) and that the initial distribution is \(\boldsymbol{\delta} = (\frac{1}{3}, \frac{1}{3}, \frac{1}{3})\)
eq_init_ns <- list(m=m,
lambda=lambda,
gamma = gamma,
delta = rep(1, m)/m)(mod3h <- pois.HMM.mle(eq_dat$Count,eq_init_ns$m,eq_init_ns$lambda,eq_init_ns$gamma,delta=eq_init_ns$delta,stationary=FALSE))## $m
## [1] 3
##
## $lambda
## [1] 13.13374 19.71312 29.70964
##
## $gamma
## [,1] [,2] [,3]
## [1,] 9.392937e-01 0.03209754 0.02860881
## [2,] 4.040106e-02 0.90643731 0.05316163
## [3,] 9.052023e-12 0.19025401 0.80974599
##
## $delta
## [1] 9.999998e-01 8.700482e-08 8.281945e-08
##
## $code
## [1] 1
##
## $mllk
## [1] 328.5275
##
## $AIC
## [1] 679.055
##
## $BIC
## [1] 708.4561Parametric Bootstrapping for Confidence Intervals
We can also perform parameteric bootstrapping to obtain confidence intervals of the above estimates.
Note: See Obtaining Standard Errors and Confidence Intervals.
Step 1: Fit the model
eq_mle <- pois.HMM.mle(eq_dat$Count,eq_init_ns$m,eq_init_ns$lambda,eq_init_ns$gamma,delta=eq_init_ns$delta,stationary=FALSE)Step 2: Generate a bootstrap sample from the fitted model.
The following function generates a sample from a Poisson-HMM of length ny from starting values m number of states, lambda means for the Poisson state-dependent distribution, and delta initial probabilities which are stored as mod (list).
pois.HMM.generate_sample <- function(ny,mod){
mvect <- 1:mod$m
state <- numeric(ny)
state[1] <- sample(mvect,1,prob=mod$delta)
for (i in 2:ny) state[i] <- sample(mvect,1,prob=mod$gamma[state[i-1],])
x <- rpois(ny,lambda=mod$lambda[state])
return(x)
}boot_sample <- pois.HMM.generate_sample(length(eq_dat$Count), eq_mle)Step 3: Estimate parameters using the bootstrap sample.
boot_mle <- pois.HMM.mle(boot_sample, eq_mle$m, eq_mle$lambda, eq_mle$gamma)## Warning in nlm(pois.HMM.mllk, parvect0, x = x, m = m, stationary = stationary):
## NA/Inf replaced by maximum positive valueStep 4: Repeat (for a large) B times.
We can combine the parts above into the pois.HMM.boot function.
pois.HMM.boot <- function(B, x, m, lambda0, gamma0, delta0=NULL, stationary=TRUE){
# Fit the model
mod <- pois.HMM.mle(x, m, lambda0, gamma0, stationary=stationary)
# Initialize
bs_sample <- list()
bs_mod <- list()
# Generate B bootstrap samples
for(i in 1:B){
a = TRUE
while(a){
tryCatch({
set.seed(i)
bs_sample[[i]] <- pois.HMM.generate_sample(length(x), mod)
bs_mod[[i]] <- pois.HMM.mle(bs_sample[[i]], m, mod$lambda, mod$gamma, mod$delta)
a = FALSE
},
error=function(w){
a = TRUE
})
}
}
return(bs_mod)
}Note: Some errors would occasionally arise from solving the stationary distribution of bootstrap samples that may not have had a unique stationary distribution. To overcome this, the tryCatch function was used to throw out these problematic samples and regenerate a new sample. As there were at most two samples that resulted in the error, the generated bootstrap samples should still be representative of the dataset.
Hence, for the three-state Poisson-HMM with parameters
\[\boldsymbol{\lambda} = (10, 20, 25)\]
\[\boldsymbol{\Gamma} = \begin{pmatrix} 0.8 & 0.1 & 0.1\\ 0.1 & 0.8 & 0.1\\ 0.1 & 0.1 & 0.8 \end{pmatrix}\]
\[\boldsymbol{\delta} = (\frac{1}{3}, \frac{1}{3}, \frac{1}{3})\]
eq_boot <- pois.HMM.boot(500, eq_dat$Count, eq_init_ns$m, eq_init_ns$lambda, eq_init_ns$gamma, delta=eq_init_ns$delta)and the \(90\%\) confidence interval using the “percentile method”
# Create dataframe for parameters from bootstrap samples
eq_lambda_boot <- sapply(eq_boot, "[[", 2)
eq_lambda_boot <- t(eq_lambda_boot)
eq_lambda_boot <- as.data.frame(eq_lambda_boot)
eq_gamma_boot <- sapply(eq_boot, "[[", 3)
eq_gamma_boot <- t(eq_gamma_boot)
eq_gamma_boot <- as.data.frame(eq_gamma_boot)
eq_delta_boot <- sapply(eq_boot, "[[", 4)
eq_delta_boot <- t(eq_delta_boot)
eq_delta_boot <- as.data.frame(eq_delta_boot)
# Obtain the 90% confidence interval
eq_boot_df <- t(cbind(sapply(eq_lambda_boot, quantile, probs=c(0.05, 0.95)),
sapply(eq_gamma_boot, quantile, probs=c(0.05, 0.95)),
sapply(eq_delta_boot, quantile, probs=c(0.05, 0.95)))) %>%
as.data.frame()
rownames(eq_boot_df)[1:3] <- paste0("lambda", 1:3)
rownames(eq_boot_df)[4:6] <- paste0("gamma", 1, 1:3)
rownames(eq_boot_df)[7:9] <- paste0("gamma", 2, 1:3)
rownames(eq_boot_df)[10:12] <- paste0("gamma", 3, 1:3)
rownames(eq_boot_df)[13:15] <- paste0("delta", 1:3)
eq_boot_df %>%
round(4) %>%
kbl(caption="Bootstrap 90% Confidence Intervals for the parameters of the three-state HMM.") %>%
kable_minimal(full_width=T)| 5% | 95% | |
|---|---|---|
| lambda1 | 11.8405 | 14.6116 |
| lambda2 | 16.9308 | 22.0249 |
| lambda3 | 19.4806 | 33.0097 |
| gamma11 | 0.8232 | 0.9880 |
| gamma12 | 0.0137 | 0.2132 |
| gamma13 | 0.0000 | 0.0000 |
| gamma21 | 0.0000 | 0.1162 |
| gamma22 | 0.6603 | 0.9637 |
| gamma23 | 0.0515 | 0.5534 |
| gamma31 | 0.0000 | 0.1108 |
| gamma32 | 0.0000 | 0.1908 |
| gamma33 | 0.4466 | 0.9485 |
| delta1 | 0.1370 | 0.7485 |
| delta2 | 0.1114 | 0.6606 |
| delta3 | 0.0404 | 0.3770 |
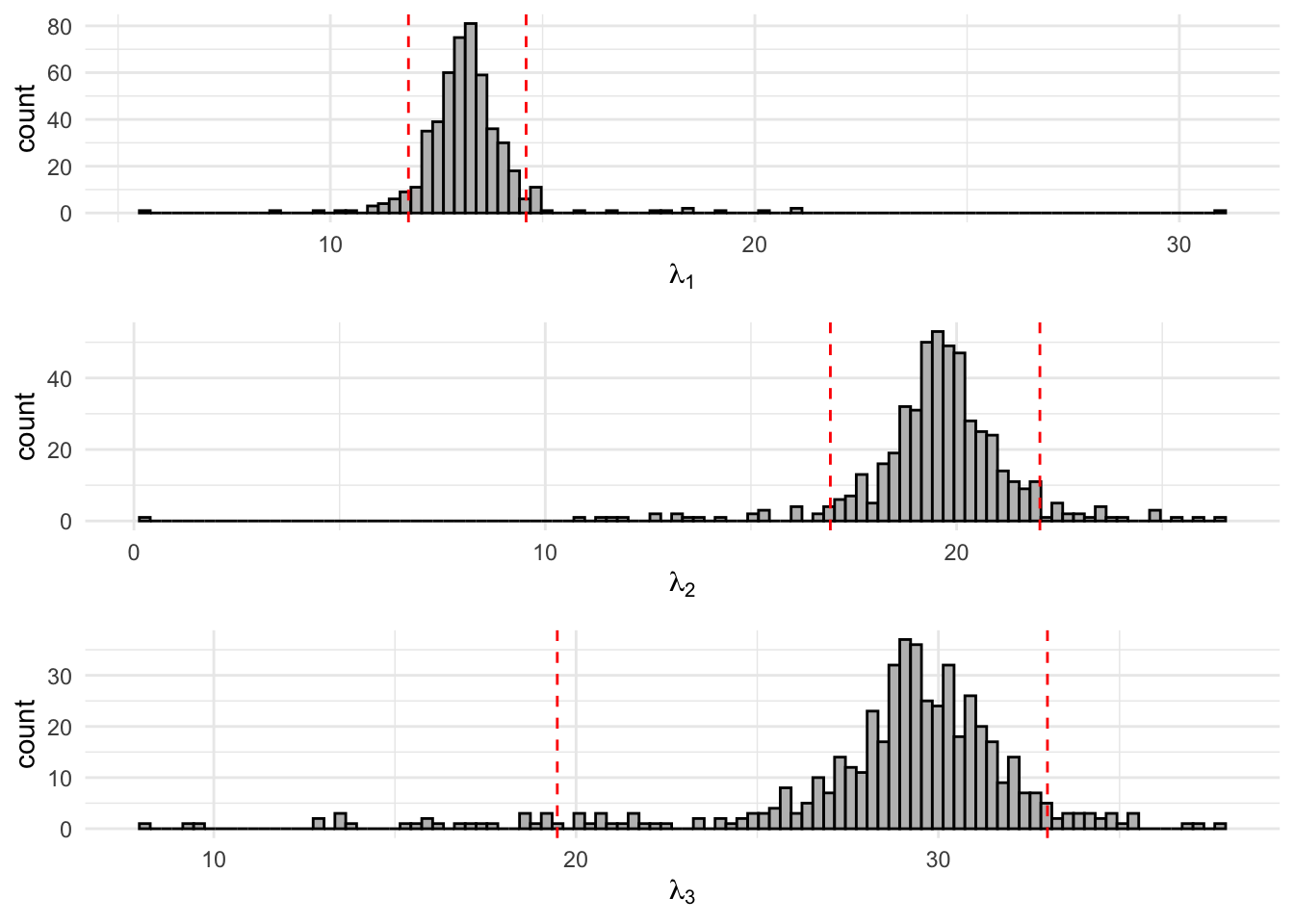
Figure 8.4: 500 Bootstrap samples for the state-dependent parameters of the fitted three-state Poisson-HMM. The area within the red dotted lines correspond to the 90% confidence interval.
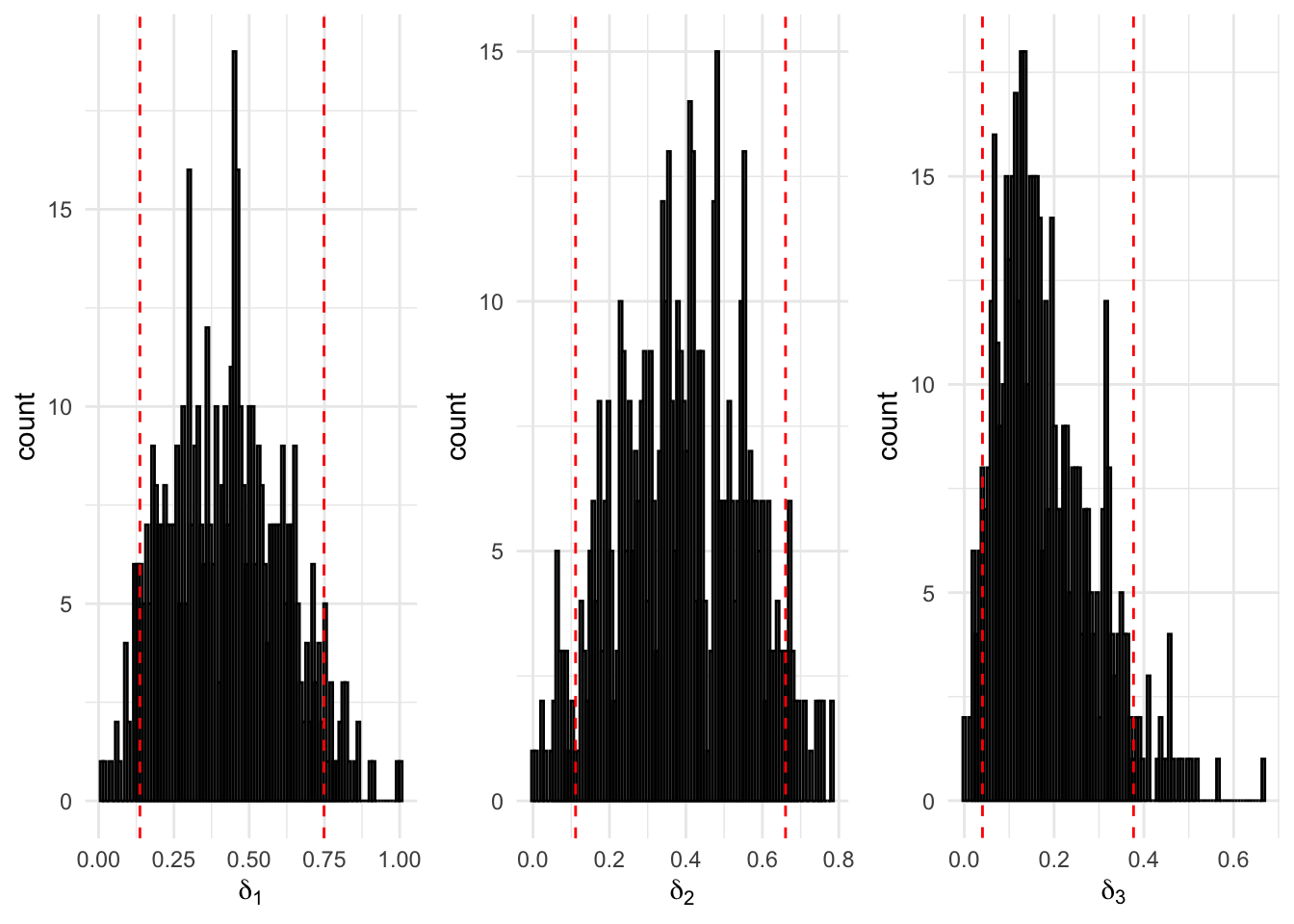
Figure 8.5: 500 Bootstrap samples for the initial probabilities of the fitted three-state Poisson-HMM. The area within the red dotted lines correspond to the 90% confidence interval.
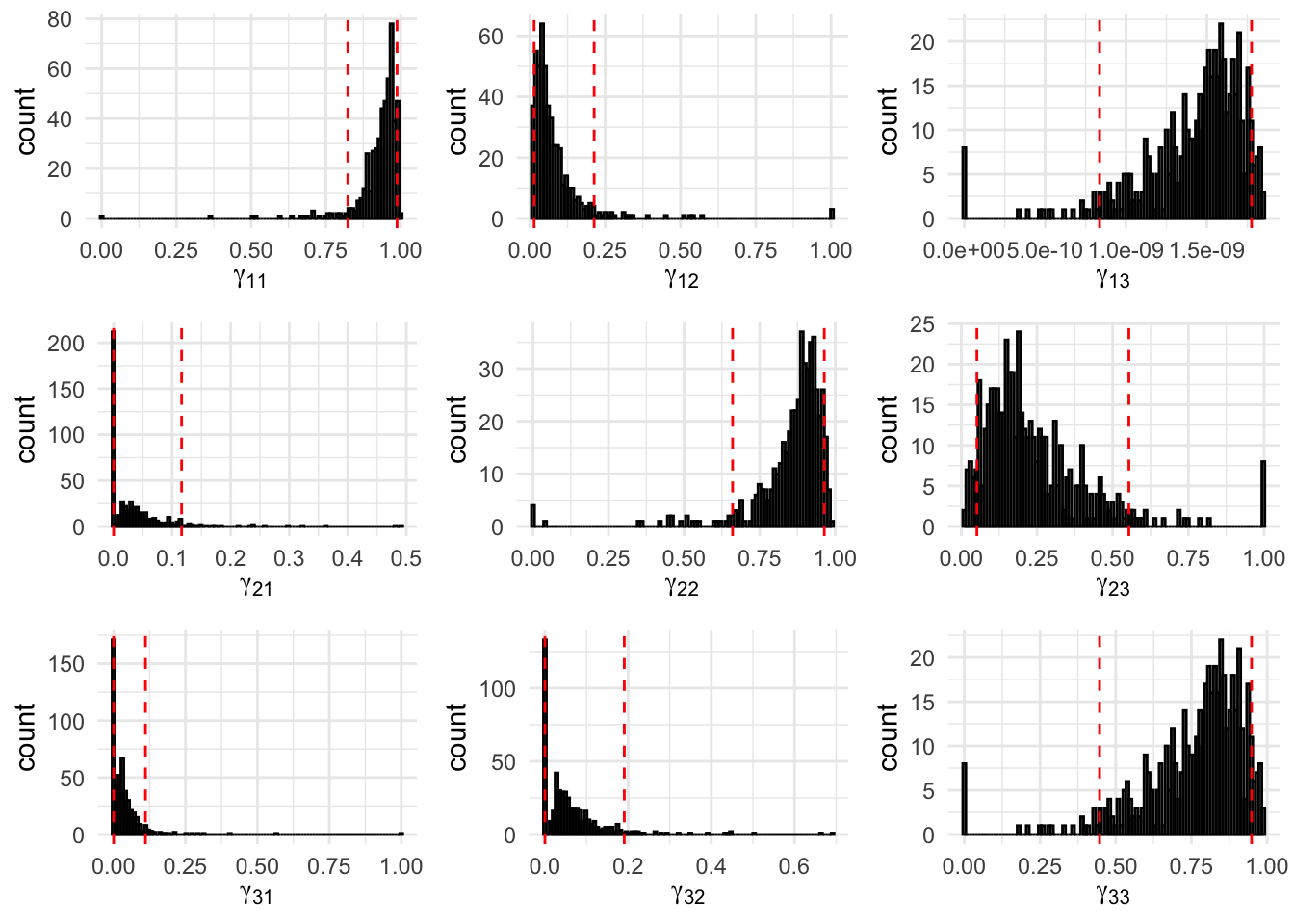
Figure 8.6: 500 Bootstrap samples for the transition probabilities of the fitted three-state Poisson-HMM. The area within the red dotted lines correspond to the 90% confidence interval.
Using momentuHMM
We can also fit the three-state Poisson-HMM from the above using momentuHMM, which is a popular package for the maximum likelihood analysis of animal movement behaviour using multivariate Hidden Markov Models. McClintock and Michelot (2018)
First, convert the data into the prepData object and set the names of the states. Since
# prepData
eq_prepped <- prepData(eq_dat,
coordNames = NULL,
covNames = c("Count"))
# Name the states
stateNames <- c("state1", "state2", "state3")Next, specify the state-dependent model. In our case, we fit the intercept only model.
# Fit intercept only model
formula = ~ 1Now, fit the model using fitHMM
mod3h_momentuHMM <- fitHMM(eq_prepped,
nbStates=3,
dist=list(Count="pois"),
Par0 = list(Count = eq_init_ns$lambda, delta=eq_init_ns$delta),
formula = formula,
stateNames = stateNames,
stationary = FALSE)The resulting MLEs are very similar to that obtained from running the functions from above.
mod3h_momentuHMM ## Value of the maximum log-likelihood: -328.5884
##
##
## Count parameters:
## -----------------
## state1 state2 state3
## lambda 13.13513 19.70822 29.70794
##
## Regression coeffs for the transition probabilities:
## ---------------------------------------------------
## 1 -> 2 1 -> 3 2 -> 1 2 -> 3 3 -> 1 3 -> 2
## (Intercept) -3.42222 -3.520977 -3.119039 -2.83615 -28.60659 -1.448253
##
## Transition probability matrix:
## ------------------------------
## state1 state2 state3
## state1 9.414330e-01 0.03072828 0.02783867
## state2 4.007763e-02 0.90674107 0.05318130
## state3 3.052488e-13 0.19027062 0.80972938
##
## Initial distribution:
## ---------------------
## state1 state2 state3
## 1.000000e+00 1.833456e-08 1.427794e-08The corresponding \(95 \%\) confidence intervals are
CIreal(mod3h_momentuHMM)## $Count
## $Count$est
## state1 state2 state3
## lambda 13.13513 19.70822 29.70794
##
## $Count$se
## state1 state2 state3
## lambda 0.6600995 0.8708107 1.679509
##
## $Count$lower
## state1 state2 state3
## lambda 11.84136 18.00146 26.41616
##
## $Count$upper
## state1 state2 state3
## lambda 14.4289 21.41497 32.99972
##
##
## $gamma
## $gamma$est
## state1 state2 state3
## state1 9.414330e-01 0.03072828 0.02783867
## state2 4.007763e-02 0.90674107 0.05318130
## state3 3.052488e-13 0.19027062 0.80972938
##
## $gamma$se
## state1 state2 state3
## state1 4.445528e-02 0.04082282 0.03293175
## state2 3.190339e-02 0.05399426 0.04414623
## state3 4.291928e-14 0.11385140 0.11385140
##
## $gamma$lower
## state1 state2 state3
## state1 7.679853e-01 0.002155143 0.002630290
## state2 8.151374e-03 0.735557101 0.009973817
## state3 2.317242e-13 0.052321545 0.499974591
##
## $gamma$upper
## state1 state2 state3
## state1 9.873516e-01 0.3175655 0.2371871
## state2 1.749874e-01 0.9714172 0.2384795
## state3 4.021022e-13 0.5000254 0.9476785
##
##
## $delta
## $delta$est
## state1 state2 state3
## ID:Animal1 1 1.833456e-08 1.427794e-08
##
## $delta$se
## state1 state2 state3
## ID:Animal1 1.046115e-13 1.046128e-13 1.493643e-21
##
## $delta$lower
## state1 state2 state3
## ID:Animal1 1 1.833436e-08 1.427794e-08
##
## $delta$upper
## state1 state2 state3
## ID:Animal1 1 1.833477e-08 1.427794e-08Fitting a Poisson-HMM by the EM Algorithm
Note: See EM Algorithm (Baum-Welch)
Alternatively, we may obtain MLEs of the three-state Poisson HMM by the EM algorithm.
E-Step Replace all quantities \(v_jk (t)\), \(u_j (t)\) by their conditional expectations given the observations \(\boldsymbol{x}^{(T)}\) and current parameter estimates:
\[\hat{u}_j = \Pr (C_t = j|\boldsymbol{x}^{(T)}) = \frac{\alpha_t (j) \beta_t (j)}{L_T}\]
\[{\hat{v}}_{jk} (t) = \Pr (C_{t-1} = j, C_t = k| \boldsymbol{x}^{(T)}) \frac{\alpha_{t-1} (j) \gamma_{jk} p_k (x_t) \beta_t (k)}{L_T}\]
This can be broken down into
Step 1: Compute the forward and backward probabilities.
We can use the same forward algorithm from the above and apply the backward algorithm pois.HMM.lbackward.
The general algorithm is:
\[\begin{align} \boldsymbol{\phi}_T \leftarrow \frac{\boldsymbol{1}}{w_T}$; $l \leftarrow \log w_T\\ \text{for } t = T-1, \dots, 1\\ \boldsymbol{v} &\leftarrow \boldsymbol{\Gamma P} (x_{t+1}) \boldsymbol{\phi_{t+1}}\\ l &\leftarrow l + \log \boldsymbol{v}\\ u &\leftarrow \boldsymbol{v 1'}\\ \boldsymbol{\phi}_t &\leftarrow \frac{\boldsymbol{v}}{u}\\ l & \leftarrow l + \log u \end{align}\]
pois.HMM.lbackward<-function(x,mod){
n <- length(x)
m <- mod$m
lbeta <- matrix(NA,m,n)
lbeta[,n] <- rep(0,m)
# For t = T
foo <- rep(1/m,m)
lscale <- log(m)
# For t = (T-1),..., 1
for (i in (n-1):1){
foo <- mod$gamma%*%(dpois(x[i+1],mod$lambda)*foo)
lbeta[,i] <- log(foo)+lscale
sumfoo <- sum(foo)
foo <- foo/sumfoo
lscale <- lscale+log(sumfoo)
}
return(lbeta)
}Note: pois.HMM.lbackward takes in the same arguments as the pois.HMM.lforward.
Step 2: Replace \(u_j(t)\) by \(\hat{u}_j (t)\) and \(v_{jk}(t)\) by \(\hat{v}_{jk} (t)\).
uhat <- function(x, mod){
n <- length(x)
m <- mod$m
# Forward probabilities
la <- pois.HMM.lforward(x, mod)
# Backward probabilities
lb <- pois.HMM.lbackward(x, mod)
c <- max(la[,n])
# L_T (with some constant amount)
llk <- c + log(sum(exp(la[,n] - c)))
# Initialize
stateprobs <- matrix(NA, ncol=n, nrow=m)
## (a_t(j) b_t (j))/L_T
for(i in 1:n){
stateprobs[,i] <- exp(la[, i] + lb[, i] - llk)
}
return(stateprobs)
}
vhat <- function(x, mod){
n <- length(x)
m <- mod$m
# Forward probabilities
la <- pois.HMM.lforward(x, mod)
# Backward probabilities
lb <- pois.HMM.lbackward(x, mod)
# tpm
lgamma <- log(mod$gamma)
c <- max(la[, n])
# L_T (with some constant amount)
llk <- c + log(sum(exp(la[, n] - c)))
v <- array(0, dim=c(m, m, n))
px <- matrix(NA, nrow=mod$m, ncol=n)
for(i in 1:n){
px[, i] <- dpois(x[i], mod$lambda)
}
lpx <- log(px)
for(i in 2:n){
for(j in 1:mod$m){
for(k in 1:mod$m){
v[j, k, i] <- exp(la[j, i-1] + lgamma[j, k] + lpx[k, i] + lb[k, i] - llk)
}
}
}
return(v)
}Note: The term c in the code is a constant added to the log-forward and log-backward probabilities to prevent numerical underflow.
M-Step Maximize the complete data log-likelihood with respect to the parameter estimates, with functions of the missing data replaced by their conditional expectations.
That is, maximize
\[\begin{align} \log \left(\Pr(\boldsymbol{x}^{(T)}, \boldsymbol{c}^{(T)}) \right) &= \sum_{j=1}^m \hat{u}_j (1) + \log \delta_j + \sum_{j=1}^m \sum_{k=1}^m \left(\sum_{t=2}^T \hat{v}_{jk} (t)\right) \log \gamma_{jk} + \sum_{j=1}^m \sum_{t=1}^T \hat{u}_j (t) \log p_j (x_t) \end{align}\]
with respect to \(\boldsymbol{\theta} = (\boldsymbol{\delta}, \boldsymbol{\Gamma}, \boldsymbol{\lambda})\).
The maximizations of the parameters are given by
E_delta <- function(uhat){
d <- uhat[, 1]
return(d)
}
E_gamma <- function(vhat, mod){
num <- apply(vhat, MARGIN=c(1, 2), FUN=sum)
denom <- rowSums(num)
g <- num/denom
return(g)
}
E_pois_lambda <- function(x,mod,uhat){
n <- length(x)
m <- mod$m
xx <- matrix(rep(x, m), nrow=m, ncol=n, byrow=TRUE)
lambda_hat <- rowSums(uhat*xx)/rowSums(uhat)
return(lambda_hat)
}We can combine the parts above into the BaumWelch function. BaumWelch performs the EM algorithm by taking initial model parameters as inputs, then repeatedly fitting until convergence or the maximum number of iterations is reached (by default maxit=50).
BaumWelch <- function(x, m, lambda0, gamma0, delta0, maxit = 50){
# Initial parameters
mod_old <- list(m = m, lambda = lambda0, gamma = gamma0, delta = delta0)
iter <- 1
while(iter < maxit){
# E-step
u_hat <- uhat(x, mod_old)
v_hat <- vhat(x, mod_old)
# M-step
delta_new <- E_delta(u_hat)
gamma_new <- E_gamma(v_hat, mod_old)
lambda_new <- E_pois_lambda(x, mod_old, u_hat)
mod_old <- list(m=m, lambda=lambda_new, gamma=gamma_new, delta=delta_new)
iter = iter + 1
}
mod <- list(m=m, lambda=lambda_new, gamma=gamma_new, delta=delta_new)
return(mod)
}Hence, for the three-state Poisson-HMM with parameters
\[\boldsymbol{\lambda} = (10, 20, 25)\]
\[\boldsymbol{\Gamma} = \begin{pmatrix} 0.8 & 0.1 & 0.1\\ 0.1 & 0.8 & 0.1\\ 0.1 & 0.1 & 0.8 \end{pmatrix}\]
\[\boldsymbol{\delta} = (\frac{1}{3}, \frac{1}{3}, \frac{1}{3})\]
# Fitting a three-state HMM
BaumWelch(eq_dat$Count, eq_init_ns$m, eq_init_ns$lambda, eq_init_ns$gamma, delta = eq_init_ns$delta)## $m
## [1] 3
##
## $lambda
## [1] 13.13376 19.71316 29.70972
##
## $gamma
## [,1] [,2] [,3]
## [1,] 9.392939e-01 0.03209847 0.02860765
## [2,] 4.040172e-02 0.90643612 0.05316216
## [3,] 2.113508e-32 0.19025585 0.80974415
##
## $delta
## [1] 1.000000e+00 3.977303e-84 2.817232e-235Forecasting, Decoding, and State Prediction
Note: See Forecasting, Decoding, and State Prediction and Appendix A of the textbook.
Forecasting
The forecast probabilities \(\Pr(X_{T+h} = x|\boldsymbol{X}^{(T)} = \boldsymbol{x}^{(T)})\) can be computed by
pois.HMM.forecast <- function(xf,h=1,x,mod)
{
n <- length(x)
nxf <- length(xf)
dxf <- matrix(0,nrow=h,ncol=nxf)
foo <- mod$delta*dpois(x[1],mod$lambda)
sumfoo <- sum(foo)
lscale <- log(sumfoo)
foo <- foo/sumfoo
for (i in 2:n)
{
foo <- foo%*%mod$gamma*dpois(x[i],mod$lambda)
sumfoo <- sum(foo)
lscale <- lscale+log(sumfoo)
foo <- foo/sumfoo
}
for (i in 1:h)
{
foo <- foo%*%mod$gamma
for (j in 1:mod$m) dxf[i,] <- dxf[i,] + foo[j]*dpois(xf,mod$lambda[j])
}
return(dxf)
}Note: pois.HMM.forecast takes in arguments xf the range of \(x\)-values for the forecast probabilities, h the forecast horizon, x data, and mod list consisting of m states, lambda means for the Poisson state-dependent distribution, and delta initial probabilities.
For example, the forecast distributions for 1 to 4 years ahead with possible values up to 45 earthquakes for the three-state Poisson-HMM (non-stationary) is
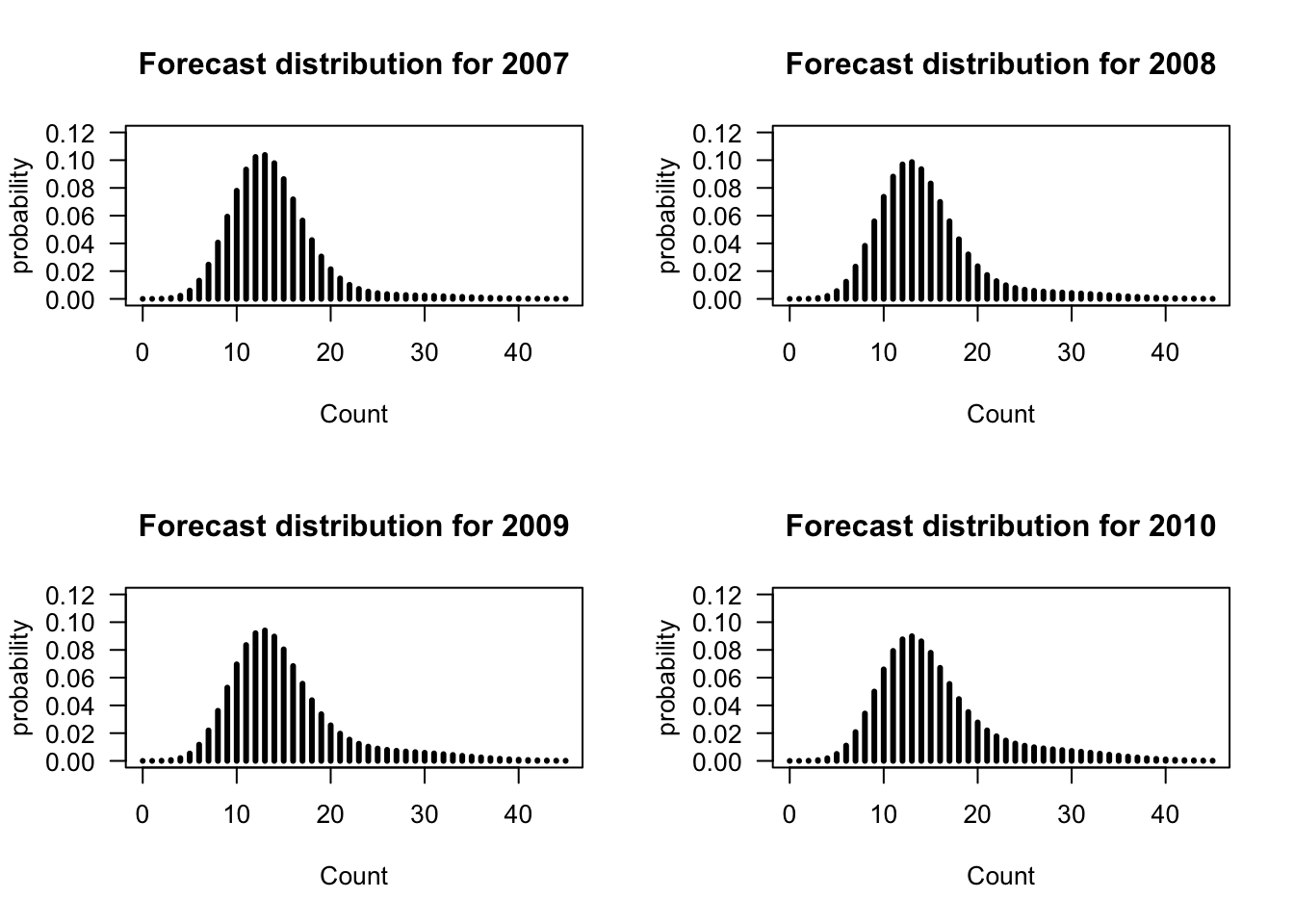
Figure 8.7: Forecast probabilities for the fitted three-state Poisson-HMM (non-stationary) for 1 to 4 years ahead.
The function from above can be modified to compute the bivariate forecast distribution.
pois.HMM.forecast_bivariate <- function(xf1, xf2, x, mod){
n <- length(x)
nxf1 <- length(xf1)
nxf2 <- length(xf2)
dxf <- matrix(0, nrow=nxf1, ncol=nxf2)
foo <- mod$delta*dpois(x[1], mod$lambda)
sumfoo <- sum(foo)
lscale <- log(sumfoo)
foo <- foo/sumfoo
for(t in 2:n){
foo <- foo%*%mod$gamma*dpois(x[t], mod$lambda)
sumfoo <- sum(foo)
lscale <- lscale + log(sumfoo)
foo <- foo/sumfoo
}
for (i in 1:nxf1){
for(j in 1:nxf2){
for(s in 1:mod$m){
dxf[i, j] <- dxf[i, j] + foo[s]*dpois(xf1[i], mod$lambda[s]) + foo[s]*dpois(xf2[j], mod$lambda[s])
}
}
}
return(foo)
}For example, \(\Pr(X_{T+1} \leq 10, X_{T+2} \leq 10|\boldsymbol{X}^{(T)})\) is
eq_forecast <- data.frame(pois.HMM.forecast_bivariate(1:10, 1:10,eq_dat$Count, eq_init_ns))
eq_forecast %>%
kbl(caption="Bivariate Forecast Distribution of $X_{T+1}$ (rows) and $X_{T+2}$ (columns).") %>%
kable_minimal(full_width=T) %>%
kable_styling()| X1 | X2 | X3 |
|---|---|---|
| 0.961864 | 0.0370039 | 0.0011321 |
Decoding
Local decoding can be computed using the forward and backward algorithms from above. Similar to computing \(\hat{u}\) in the E step of the EM algorithm, the pois.HMM.state_probs computes the state probabilities then pois.HMM.local_decoding determines the most likely state at each time.
pois.HMM.state_probs <- function(x,mod)
{
n <- length(x)
la <- pois.HMM.lforward(x,mod)
lb <- pois.HMM.lbackward(x,mod)
c <- max(la[,n])
llk <- c+log(sum(exp(la[,n]-c)))
stateprobs <- matrix(NA,ncol=n,nrow=mod$m)
for (i in 1:n) stateprobs[,i]<-exp(la[,i]+lb[,i]-llk)
return(stateprobs)
}
pois.HMM.local_decoding <- function(x,mod)
{
n <- length(x)
stateprobs <- pois.HMM.state_probs(x,mod)
ild <- rep(NA,n)
for (i in 1:n) ild[i]<-which.max(stateprobs[,i])
ild
}For example, local decoding for the three state Poisson-HMM (non-stationary) is
# Local decoding
(earthquake_local <- pois.HMM.local_decoding(eq_dat$Count, eq_init_ns))## [1] 1 1 1 1 1 3 3 3 3 3 3 3 3 3 3 2 2 2 2 1 1 1 1 2 2 2 2 2 2 2 2 2 2 2 2 2 2
## [38] 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 2 2 2 2 2 3 1 2 2 2 2 2 2 2 2 2 3 3 3 3 2 2
## [75] 2 2 2 2 2 2 2 1 1 1 1 1 1 1 1 1 2 2 1 1 1 2 2 2 2 2 2 2 2 2 1 1 1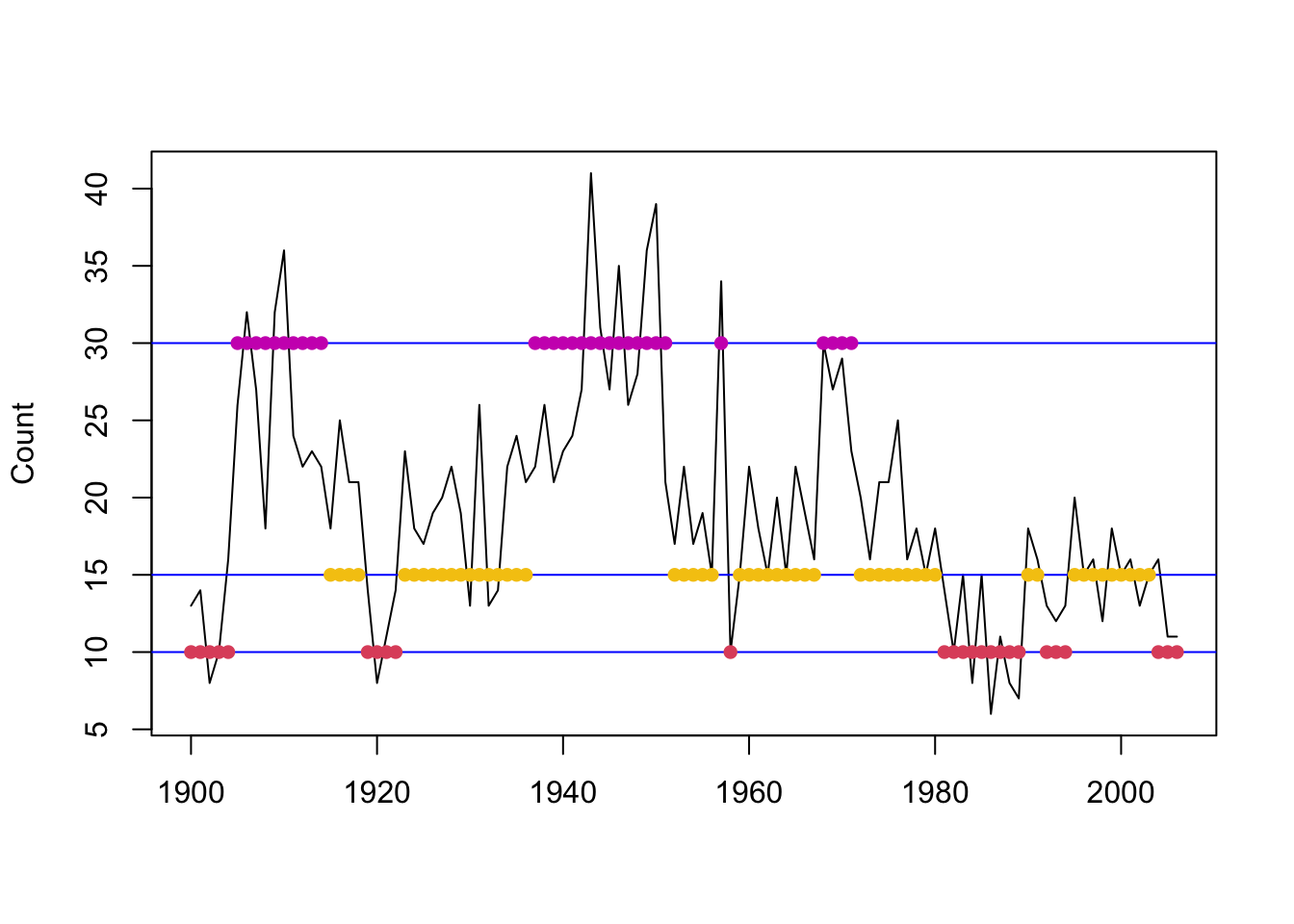
Figure 8.8: Local decoding for the three-state Poisson-HMM (non-stationary). The horizontal lines indicate the state-dependent means and the points indicate the decoded state at the given time.
We can also perform local decoding using momentuHMM
# Local decoding
## Compute state probabilities for each time step
state_probs <- stateProbs(mod3h_momentuHMM)
## Select state that maximizes probability
local_vec <- rep(NA,length(eq_dat$Count))
for(i in 1:length(eq_dat$Count)) local_vec[i] <- which.max(state_probs[i,])
local_vec## [1] 1 1 1 1 1 3 3 3 3 3 3 3 2 2 2 2 2 2 2 1 1 1 1 2 2 2 2 2 2 2 2 2 2 2 2 2 2
## [38] 2 2 2 2 3 3 3 3 3 3 3 3 3 3 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 3 3 3 2 2 2
## [75] 2 2 2 2 2 2 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1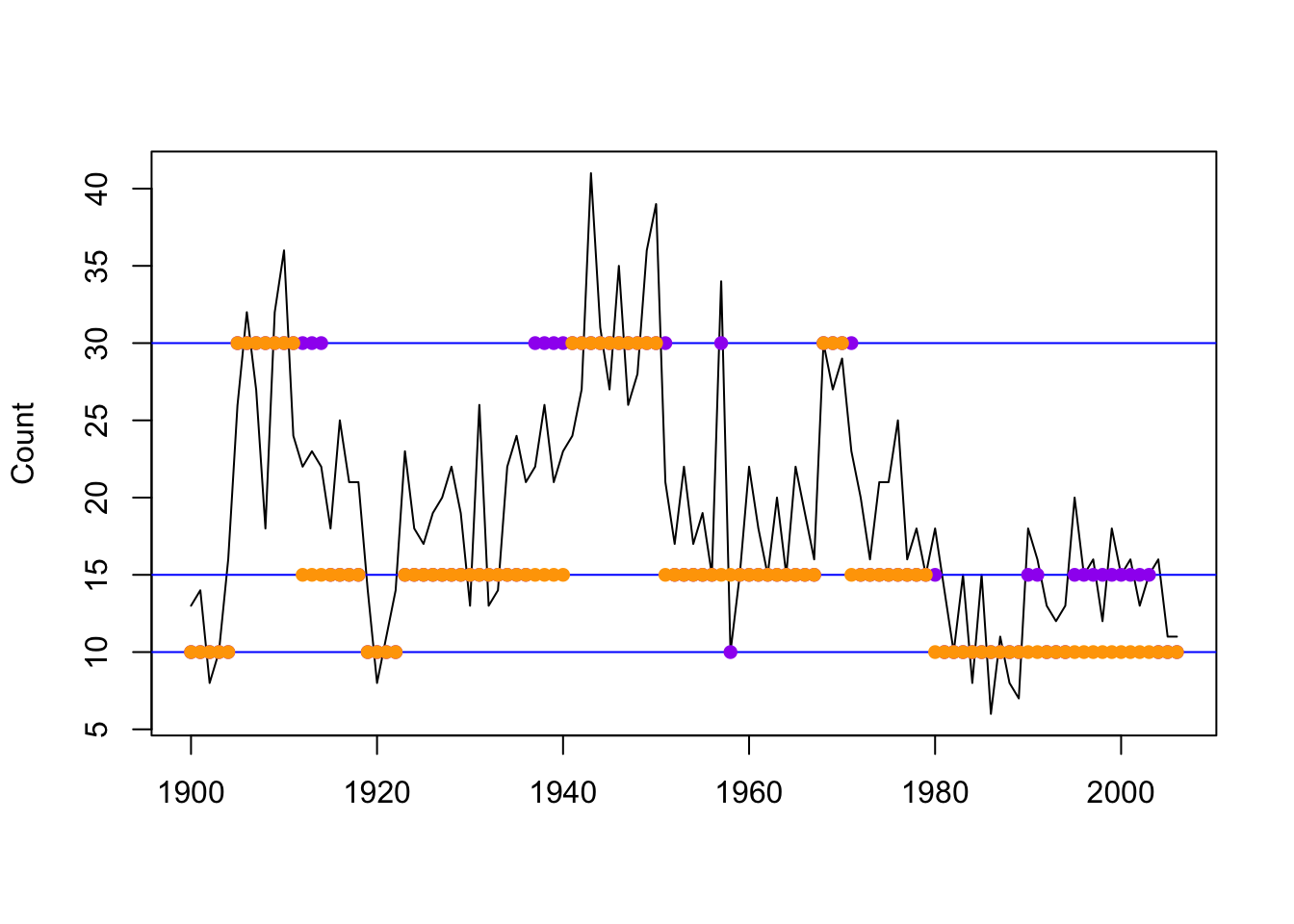
Figure 8.9: Local decoding for the three-state Poisson-HMM (non-stationary). The horizontal lines indicate the state-dependent means and the points indicate the decoded state at the given time using pois.HMM.local_decoding (purple) and momentuHMM (orange).
Global decoding can be computed using the Viterbi algorithm
pois.HMM.viterbi<-function(x,mod)
{
n <- length(x)
xi <- matrix(0,n,mod$m)
foo <- mod$delta*dpois(x[1],mod$lambda)
xi[1,] <- foo/sum(foo)
for (i in 2:n)
{
foo<-apply(xi[i-1,]*mod$gamma,2,max)*dpois(x[i],mod$lambda)
xi[i,] <- foo/sum(foo)
}
iv<-numeric(n)
iv[n] <-which.max(xi[n,])
for (i in (n-1):1)
iv[i] <- which.max(mod$gamma[,iv[i+1]]*xi[i,])
return(iv)
}For example, global decoding for the three state Poisson-HMM (non-stationary) is
(earthquake_global <- pois.HMM.viterbi(eq_dat$Count, eq_init_ns))## [1] 1 1 1 1 1 3 3 3 3 3 3 3 3 3 3 3 3 3 3 1 1 1 1 2 2 2 2 2 2 2 2 2 2 2 2 2 2
## [38] 2 3 3 3 3 3 3 3 3 3 3 3 3 3 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 3 3 3 3 2 2
## [75] 2 2 2 2 2 2 2 1 1 1 1 1 1 1 1 1 1 1 1 1 1 2 2 2 2 2 2 2 2 2 2 1 1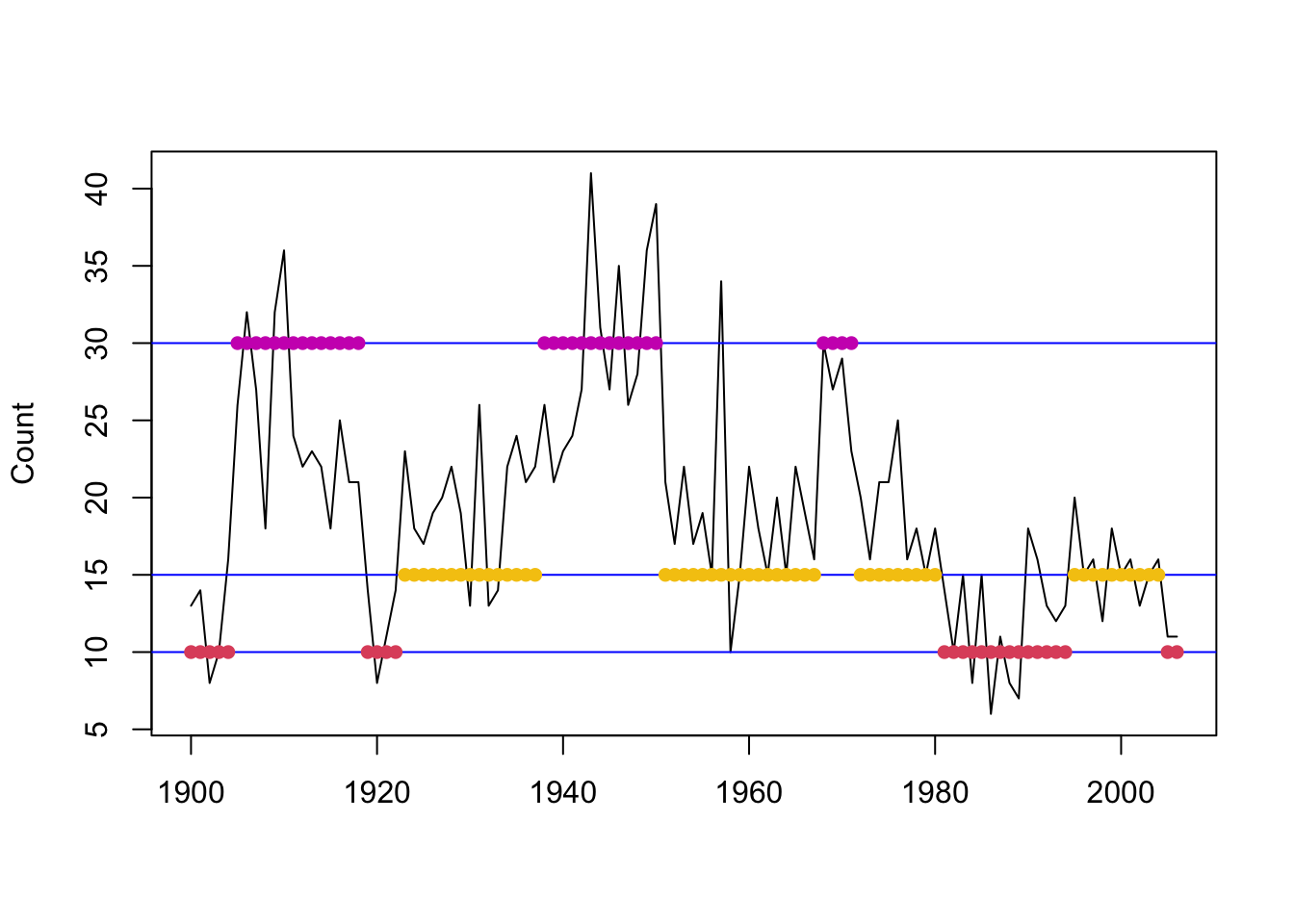
Figure 8.10: Global decoding for the three-state Poisson-HMM (non-stationary).
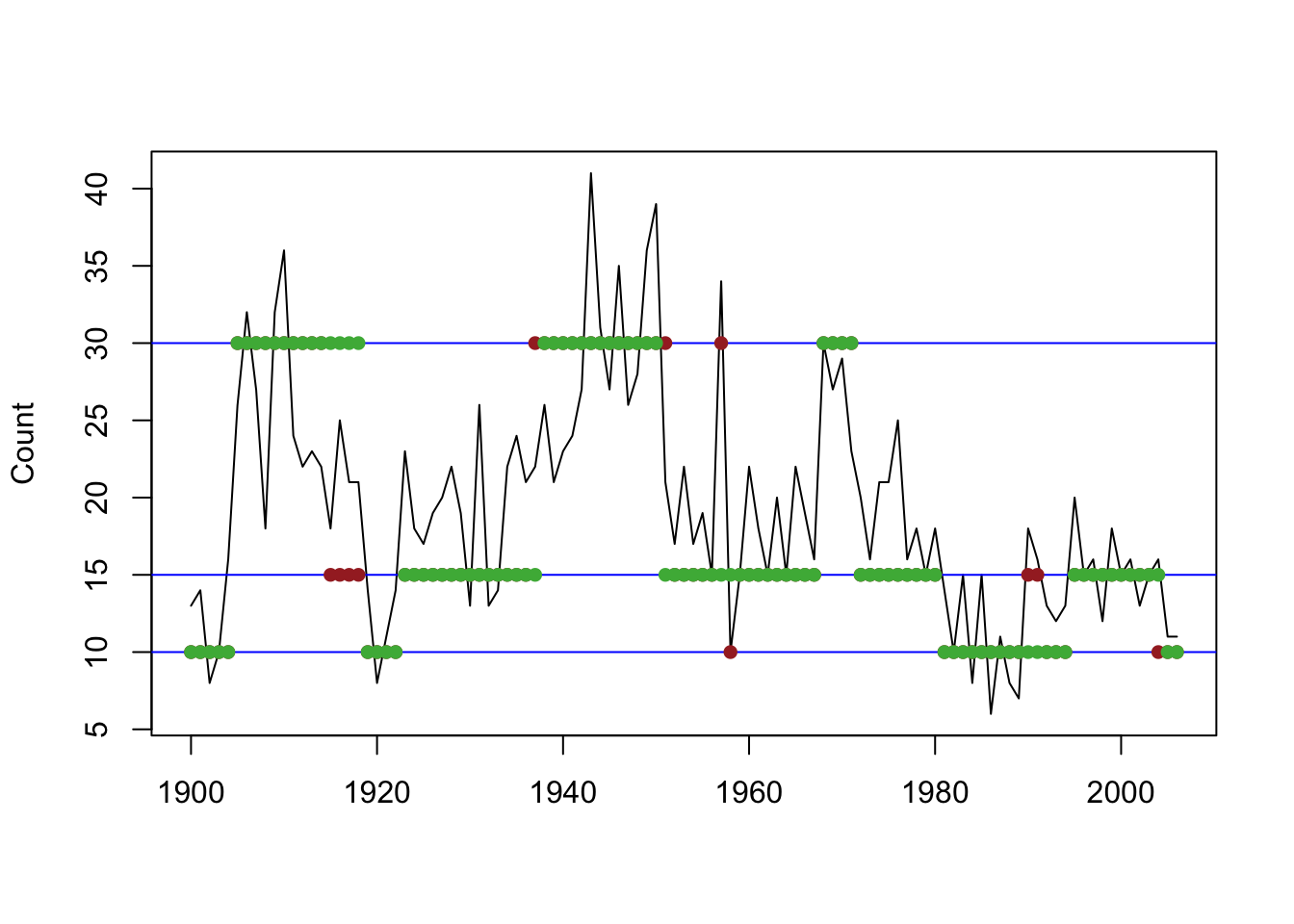
Figure 8.11: Global (green) and local (brown) decoding for the three-state Poisson-HMM (non-stationary).
The result of local and global decoding are very similar but not identical.
Similarly, we can perform global decoding using momentuHMM
(global_vec <- viterbi(mod3h_momentuHMM))## [1] 1 1 1 1 1 3 3 3 3 3 3 2 2 2 2 2 2 2 2 1 1 1 1 2 2 2 2 2 2 2 2 2 2 2 2 2 2
## [38] 2 2 2 2 2 3 3 3 3 3 3 3 3 3 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 3 3 3 2 2 2
## [75] 2 2 2 2 2 2 2 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1plotStates(mod3h_momentuHMM)## Decoding state sequence... DONE
## Computing state probabilities... DONE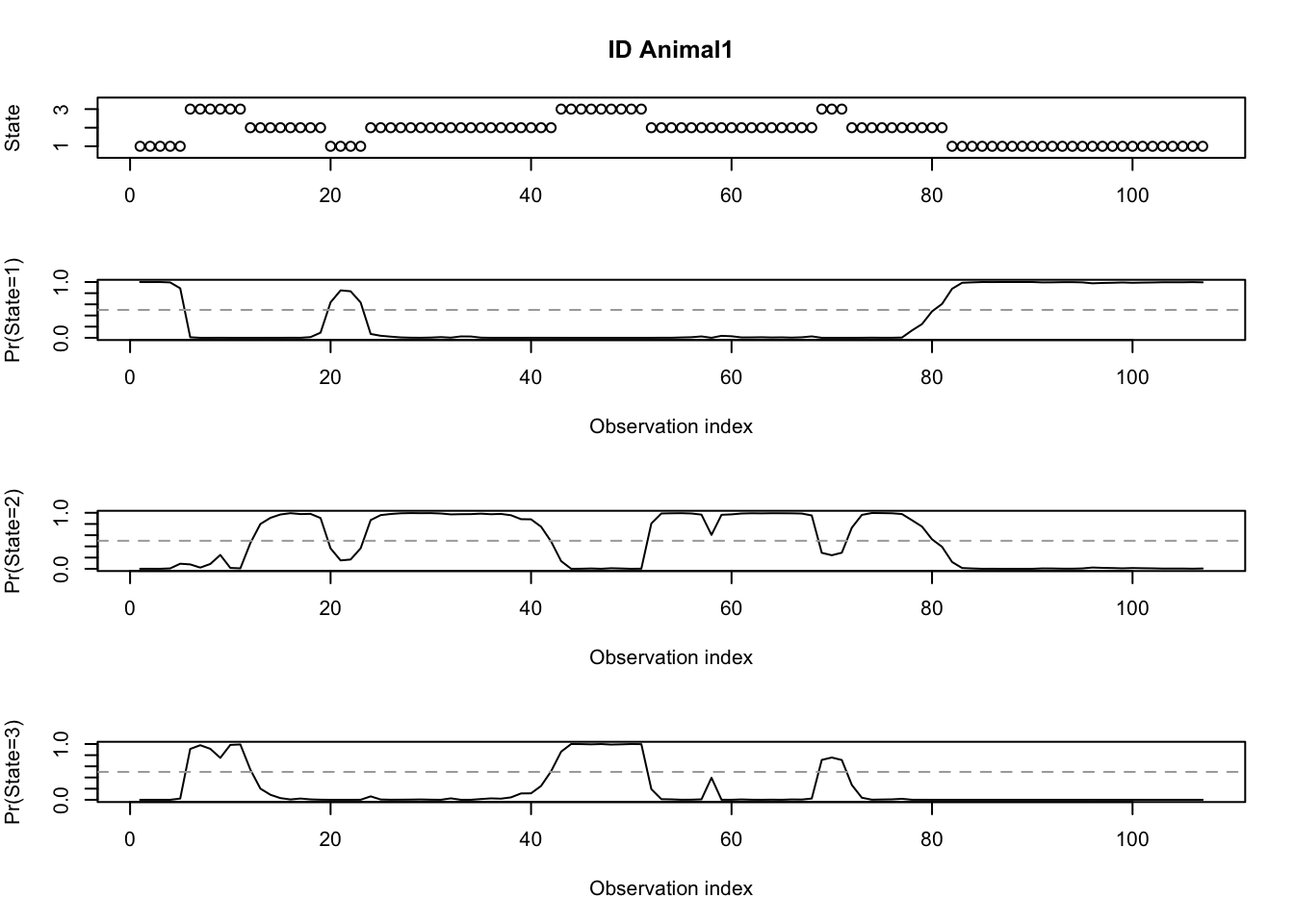
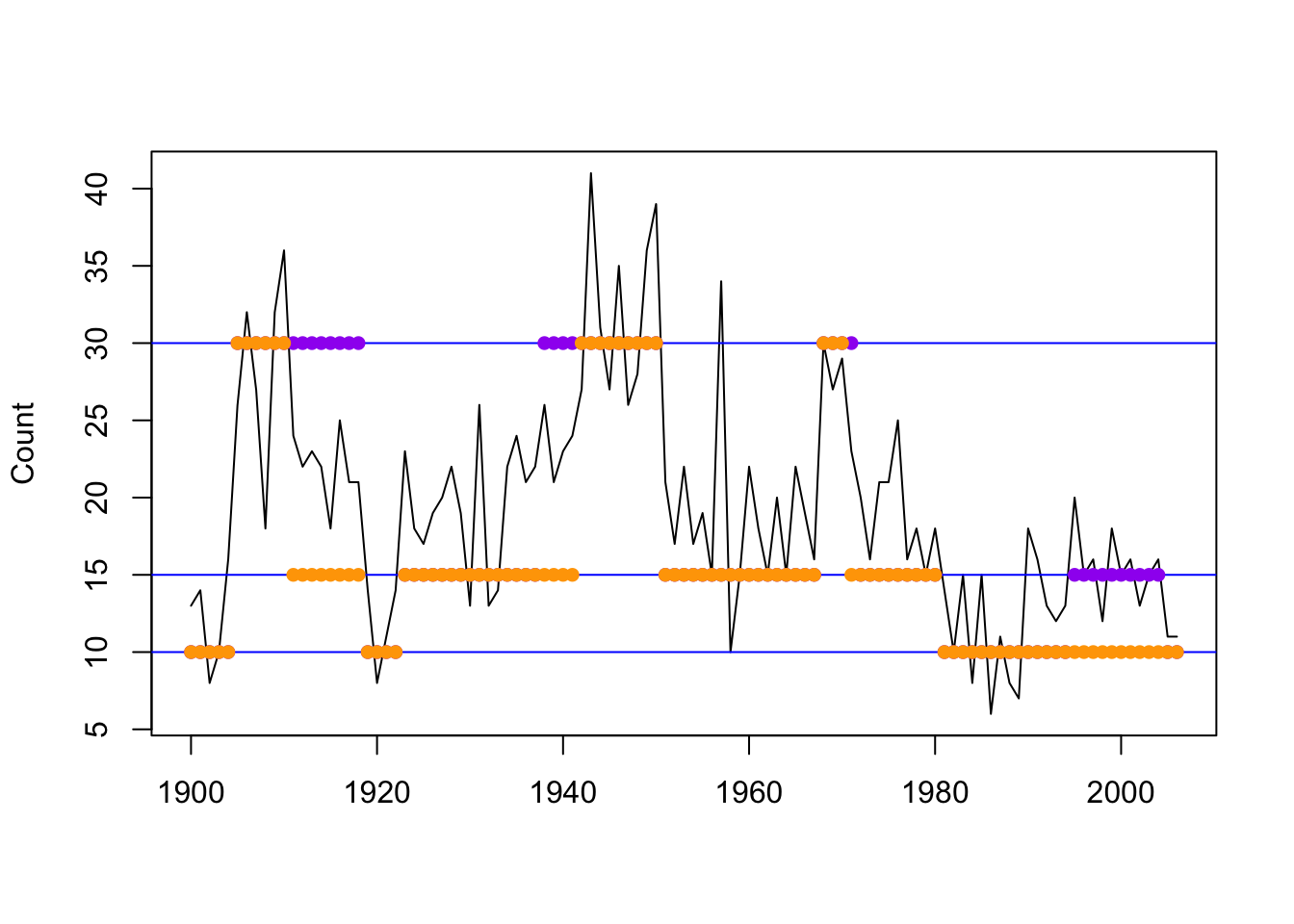
Figure 8.12: Global decoding for the three-state Poisson-HMM using pois.HMM.gocal_decoding (purple) and momentuHMM (orange).
There appears to be some differences between the results obtained from our function and from momentuHMM.
State Predictions
The state prediction for \(h\) steps ahead can be computed by
pois.HMM.state_prediction <- function(h=1,x,mod)
{
n <- length(x)
la <- pois.HMM.lforward(x,mod)
c <- max(la[,n])
llk <- c+log(sum(exp(la[,n]-c)))
statepreds <- matrix(NA,ncol=h,nrow=mod$m)
foo <- exp(la[,n]-llk)
for (i in 1:h){
foo<-foo%*%mod$gamma
statepreds[,i]<-foo
}
return(statepreds)
}For example, the state predictions for \(h=1, \dots, 5\) for the three-state Poisson-HMM (non-stationary)
pois.HMM.state_prediction(h=5, eq_dat$Count, eq_init_ns)## [,1] [,2] [,3] [,4] [,5]
## [1,] 0.7733048 0.6413134 0.5489194 0.4842436 0.4389705
## [2,] 0.1259027 0.1881319 0.2316923 0.2621846 0.2835292
## [3,] 0.1007924 0.1705547 0.2193883 0.2535718 0.2775003The stationary distribution is
# Compute stationary distribution
solve(t(diag(m)-gamma+1),rep(1,m))## [1] 0.3333333 0.3333333 0.3333333Notice that as \(h \rightarrow \infty\), the state distribution approaches the stationary distribution.
Fitting a Poisson-HMM using STAN
Note: See Bayesian Inference for HMMs, Leos-Barajas and Michelot (2018), and Damiano, Peterson, and Weylandt (2017).
We can perform Bayesian inference for the three-state Poisson-HMM using STAN Stan Development Team (2023).
In STAN, the main building blocks are the:
data block: used to define the input data for the model to fit in STAN. Declare the data types, their dimensions, any restrictions, and their names of the data to model.
Here, we specify the length of the sequence of the earthquake series, the number of states, and the vector of observed earthquake counts.
'data{
int<lower=0> m; // number of states
int<lower=1> T; // length of sequence
int<lower=0> x[T]; // observations
}
'parameters block: used to define the parameters of the model that will be estimated. Declare the data types, their dimensions, any restrictions, and their names of the parameters to model.
Here, we specify the state-dependent means, transition probability matrix, and the initial distribution (assuming non-stationarity).
'parameters{
simplex[m] Gamma[m]; // transition probability matrix
positive_ordered[m] lambda; // state-dependent means (ordered to prevent label switching)
}transformed parameters block: used to define any derived parameters that are not directly estimated but are derived from the parameters defined in the previous block. Declare the data types, their dimensions, any restrictions, their names, and the operation of the parameters to transform.
If we assume that the Markov Chain is stationary, then we can compute the stationary distribution from the tpm.
'transformed parameters{
matrix[m, m] ta; // tpm used to compute stationary distribution
simplex[m] statdist; // stationary distribution
for(j in 1:m){
for(i in 1:m){
ta[i, j] = Gamma[i, j];
}
}
//compute stationary distribution
statdist = to_vector((to_row_vector(rep_vector(1.0, m))/
(diag_matrix(rep_vector(1.0, m)) - ta + rep_matrix(1, m, m))));
}
'Note: The tpm is transposed so that each row corresponds to the probabilities of transitioning into a state, rather than out of a state.
model block: used to define the prior distribution, likelihood, and any other necessary variables.
Here, we use the following prior specifications:
\[\boldsymbol{\lambda} \sim Gamma(\alpha=1, \beta=0.01)\]
\[\boldsymbol{\Gamma} \sim Unif(-\infty, \infty)\]
\[\boldsymbol{\delta} \sim Unif(-\infty, \infty)\] The forward algorithm can be used to obtain the posterior distribution of the hidden states at a given time \(t\) based on all the information available up to that time \(\Pr(C_t|\boldsymbol{X}^{(t)})\).
\[\begin{align} \log (\alpha_t, j) &= \log \left(\sum_{i=1}^m \gamma_{ij} \alpha_{t-1, i} \right)\\ &= \log \left(\sum_{i=1}^m \exp (\log (\gamma_{ij}) + \log (\alpha_{t-1, i}))\right) \end{align}\]
'model{
vector[m] log_Gamma_tr[m];
vector[m] lp;
vector[m] lp_p1;
// priors
lambda ~ gamma(1, 0.01);
// transpose tpm and take log of entries
for(i in 1:m)
for(j in 1:m)
log_Gamma_tr[j, i] = log(Gamma[i, j]);
// forward algorithm
// for t = 1
for(i in 1:m)
lp[i] = log(statdist[i]) + poisson_lpmf(x[1]|lambda[i]);
// for t > 1
for(t in 2:T){
for (i in 1:m)
lp_p1[i] = log_sum_exp(log_Gamma_tr[i] + lp) + poisson_lpmf(x[t]|lambda[i]);
lp = lp_p1;
}
target += log_sum_exp(lp);
}
'## [1] "model{\n vector[m] log_Gamma_tr[m];\n vector[m] lp;\n vector[m] lp_p1;\n\n // priors\n lambda ~ gamma(1, 0.01);\n\n // transpose tpm and take log of entries\n for(i in 1:m)\n for(j in 1:m)\n log_Gamma_tr[j, i] = log(Gamma[i, j]);\n \n // forward algorithm\n // for t = 1\n for(i in 1:m)\n lp[i] = log(statdist[i]) + poisson_lpmf(x[1]|lambda[i]);\n \n // for t > 1\n for(t in 2:T){\n for (i in 1:m)\n lp_p1[i] = log_sum_exp(log_Gamma_tr[i] + lp) + poisson_lpmf(x[t]|lambda[i]);\n \n lp = lp_p1;\n }\n target += log_sum_exp(lp);\n }\n"All together,
pois.HMM.stan <-
'data{
int<lower=0> m; // number of states
int<lower=1> T; // length of sequence
int<lower=0> x[T]; // observations
}
parameters{
simplex[m] Gamma[m]; // tpm
positive_ordered[m] lambda; // ordered mean of state-dependent (prevent label switching)
}
transformed parameters{
matrix[m, m] ta; // tpm used to compute stationary distribution
simplex[m] statdist; // stationary distribution
for(j in 1:m){
for(i in 1:m){
ta[i, j] = Gamma[i, j];
}
}
//compute stationary distribution
statdist = to_vector((to_row_vector(rep_vector(1.0, m))/
(diag_matrix(rep_vector(1.0, m)) - ta + rep_matrix(1, m, m))));
}
model{
// initialise
vector[m] log_Gamma_tr[m];
vector[m] lp;
vector[m] lp_p1;
// priors
lambda ~ gamma(1, 0.01);
// transpose tpm and take log of entries
for(i in 1:m)
for(j in 1:m)
log_Gamma_tr[j, i] = log(Gamma[i, j]);
// forward algorithm
// for t = 1
for(i in 1:m)
lp[i] = log(statdist[i]) + poisson_lpmf(x[1]|lambda[i]);
// loop over observations
for(t in 2:T){
// loop over states
for (i in 1:m)
lp_p1[i] = log_sum_exp(log_Gamma_tr[i] + lp) + poisson_lpmf(x[t]|lambda[i]);
lp = lp_p1;
}
target += log_sum_exp(lp);
}
'Then for the three state-Poisson HMM,
# Format data into list for STAN
stan_data <- list(T=dim(eq_dat)[1], m=3, x=eq_dat$Count)Running 2000 iterations for each of the 4 chains, with the first 1000 draws drawn during the warm-up phase,
pois.HMM.stanfit <- stan(model_code = pois.HMM.stan, data = stan_data, refresh=2000)## Trying to compile a simple C file## Running /Library/Frameworks/R.framework/Resources/bin/R CMD SHLIB foo.c
## clang -arch arm64 -I"/Library/Frameworks/R.framework/Resources/include" -DNDEBUG -I"/Library/Frameworks/R.framework/Versions/4.2-arm64/Resources/library/Rcpp/include/" -I"/Library/Frameworks/R.framework/Versions/4.2-arm64/Resources/library/RcppEigen/include/" -I"/Library/Frameworks/R.framework/Versions/4.2-arm64/Resources/library/RcppEigen/include/unsupported" -I"/Library/Frameworks/R.framework/Versions/4.2-arm64/Resources/library/BH/include" -I"/Library/Frameworks/R.framework/Versions/4.2-arm64/Resources/library/StanHeaders/include/src/" -I"/Library/Frameworks/R.framework/Versions/4.2-arm64/Resources/library/StanHeaders/include/" -I"/Library/Frameworks/R.framework/Versions/4.2-arm64/Resources/library/RcppParallel/include/" -I"/Library/Frameworks/R.framework/Versions/4.2-arm64/Resources/library/rstan/include" -DEIGEN_NO_DEBUG -DBOOST_DISABLE_ASSERTS -DBOOST_PENDING_INTEGER_LOG2_HPP -DSTAN_THREADS -DBOOST_NO_AUTO_PTR -include '/Library/Frameworks/R.framework/Versions/4.2-arm64/Resources/library/StanHeaders/include/stan/math/prim/mat/fun/Eigen.hpp' -D_REENTRANT -DRCPP_PARALLEL_USE_TBB=1 -I/opt/R/arm64/include -fPIC -falign-functions=64 -Wall -g -O2 -c foo.c -o foo.o
## In file included from <built-in>:1:
## In file included from /Library/Frameworks/R.framework/Versions/4.2-arm64/Resources/library/StanHeaders/include/stan/math/prim/mat/fun/Eigen.hpp:13:
## In file included from /Library/Frameworks/R.framework/Versions/4.2-arm64/Resources/library/RcppEigen/include/Eigen/Dense:1:
## In file included from /Library/Frameworks/R.framework/Versions/4.2-arm64/Resources/library/RcppEigen/include/Eigen/Core:88:
## /Library/Frameworks/R.framework/Versions/4.2-arm64/Resources/library/RcppEigen/include/Eigen/src/Core/util/Macros.h:628:1: error: unknown type name 'namespace'
## namespace Eigen {
## ^
## /Library/Frameworks/R.framework/Versions/4.2-arm64/Resources/library/RcppEigen/include/Eigen/src/Core/util/Macros.h:628:16: error: expected ';' after top level declarator
## namespace Eigen {
## ^
## ;
## In file included from <built-in>:1:
## In file included from /Library/Frameworks/R.framework/Versions/4.2-arm64/Resources/library/StanHeaders/include/stan/math/prim/mat/fun/Eigen.hpp:13:
## In file included from /Library/Frameworks/R.framework/Versions/4.2-arm64/Resources/library/RcppEigen/include/Eigen/Dense:1:
## /Library/Frameworks/R.framework/Versions/4.2-arm64/Resources/library/RcppEigen/include/Eigen/Core:96:10: fatal error: 'complex' file not found
## #include <complex>
## ^~~~~~~~~
## 3 errors generated.
## make: *** [foo.o] Error 1
##
## SAMPLING FOR MODEL '9875485ac4343cdf02f82dbe9761d256' NOW (CHAIN 1).
## Chain 1:
## Chain 1: Gradient evaluation took 0.000123 seconds
## Chain 1: 1000 transitions using 10 leapfrog steps per transition would take 1.23 seconds.
## Chain 1: Adjust your expectations accordingly!
## Chain 1:
## Chain 1:
## Chain 1: Iteration: 1 / 2000 [ 0%] (Warmup)
## Chain 1: Iteration: 1001 / 2000 [ 50%] (Sampling)
## Chain 1: Iteration: 2000 / 2000 [100%] (Sampling)
## Chain 1:
## Chain 1: Elapsed Time: 0.820039 seconds (Warm-up)
## Chain 1: 0.450072 seconds (Sampling)
## Chain 1: 1.27011 seconds (Total)
## Chain 1:
##
## SAMPLING FOR MODEL '9875485ac4343cdf02f82dbe9761d256' NOW (CHAIN 2).
## Chain 2:
## Chain 2: Gradient evaluation took 5e-05 seconds
## Chain 2: 1000 transitions using 10 leapfrog steps per transition would take 0.5 seconds.
## Chain 2: Adjust your expectations accordingly!
## Chain 2:
## Chain 2:
## Chain 2: Iteration: 1 / 2000 [ 0%] (Warmup)
## Chain 2: Iteration: 1001 / 2000 [ 50%] (Sampling)
## Chain 2: Iteration: 2000 / 2000 [100%] (Sampling)
## Chain 2:
## Chain 2: Elapsed Time: 0.823041 seconds (Warm-up)
## Chain 2: 0.527402 seconds (Sampling)
## Chain 2: 1.35044 seconds (Total)
## Chain 2:
##
## SAMPLING FOR MODEL '9875485ac4343cdf02f82dbe9761d256' NOW (CHAIN 3).
## Chain 3:
## Chain 3: Gradient evaluation took 4.9e-05 seconds
## Chain 3: 1000 transitions using 10 leapfrog steps per transition would take 0.49 seconds.
## Chain 3: Adjust your expectations accordingly!
## Chain 3:
## Chain 3:
## Chain 3: Iteration: 1 / 2000 [ 0%] (Warmup)
## Chain 3: Iteration: 1001 / 2000 [ 50%] (Sampling)
## Chain 3: Iteration: 2000 / 2000 [100%] (Sampling)
## Chain 3:
## Chain 3: Elapsed Time: 0.812108 seconds (Warm-up)
## Chain 3: 0.457366 seconds (Sampling)
## Chain 3: 1.26947 seconds (Total)
## Chain 3:
##
## SAMPLING FOR MODEL '9875485ac4343cdf02f82dbe9761d256' NOW (CHAIN 4).
## Chain 4:
## Chain 4: Gradient evaluation took 4.9e-05 seconds
## Chain 4: 1000 transitions using 10 leapfrog steps per transition would take 0.49 seconds.
## Chain 4: Adjust your expectations accordingly!
## Chain 4:
## Chain 4:
## Chain 4: Iteration: 1 / 2000 [ 0%] (Warmup)
## Chain 4: Iteration: 1001 / 2000 [ 50%] (Sampling)
## Chain 4: Iteration: 2000 / 2000 [100%] (Sampling)
## Chain 4:
## Chain 4: Elapsed Time: 0.76585 seconds (Warm-up)
## Chain 4: 0.455552 seconds (Sampling)
## Chain 4: 1.2214 seconds (Total)
## Chain 4:we obtain the following posterior estimates
print(pois.HMM.stanfit,digits_summary = 3)## Inference for Stan model: 9875485ac4343cdf02f82dbe9761d256.
## 4 chains, each with iter=2000; warmup=1000; thin=1;
## post-warmup draws per chain=1000, total post-warmup draws=4000.
##
## mean se_mean sd 2.5% 25% 50% 75% 97.5%
## Gamma[1,1] 0.862 0.002 0.085 0.669 0.825 0.879 0.919 0.966
## Gamma[1,2] 0.091 0.002 0.076 0.006 0.039 0.073 0.122 0.274
## Gamma[1,3] 0.047 0.001 0.042 0.002 0.017 0.036 0.066 0.152
## Gamma[2,1] 0.090 0.002 0.065 0.014 0.048 0.077 0.115 0.237
## Gamma[2,2] 0.817 0.002 0.085 0.626 0.778 0.831 0.874 0.935
## Gamma[2,3] 0.093 0.001 0.058 0.015 0.051 0.083 0.120 0.237
## Gamma[3,1] 0.077 0.001 0.069 0.002 0.025 0.059 0.110 0.249
## Gamma[3,2] 0.231 0.002 0.115 0.055 0.148 0.214 0.300 0.493
## Gamma[3,3] 0.691 0.002 0.124 0.423 0.609 0.706 0.783 0.891
## lambda[1] 13.198 0.019 0.857 11.423 12.686 13.221 13.729 14.875
## lambda[2] 19.828 0.026 1.069 17.689 19.181 19.836 20.478 21.929
## lambda[3] 29.905 0.036 1.905 26.419 28.600 29.855 31.105 33.882
## ta[1,1] 0.862 0.002 0.085 0.669 0.825 0.879 0.919 0.966
## ta[1,2] 0.091 0.002 0.076 0.006 0.039 0.073 0.122 0.274
## ta[1,3] 0.047 0.001 0.042 0.002 0.017 0.036 0.066 0.152
## ta[2,1] 0.090 0.002 0.065 0.014 0.048 0.077 0.115 0.237
## ta[2,2] 0.817 0.002 0.085 0.626 0.778 0.831 0.874 0.935
## ta[2,3] 0.093 0.001 0.058 0.015 0.051 0.083 0.120 0.237
## ta[3,1] 0.077 0.001 0.069 0.002 0.025 0.059 0.110 0.249
## ta[3,2] 0.231 0.002 0.115 0.055 0.148 0.214 0.300 0.493
## ta[3,3] 0.691 0.002 0.124 0.423 0.609 0.706 0.783 0.891
## statdist[1] 0.391 0.002 0.142 0.143 0.286 0.380 0.485 0.688
## statdist[2] 0.418 0.002 0.130 0.172 0.326 0.416 0.509 0.679
## statdist[3] 0.191 0.001 0.092 0.056 0.124 0.177 0.240 0.414
## lp__ -345.917 0.079 2.515 -351.814 -347.287 -345.502 -344.077 -342.235
## n_eff Rhat
## Gamma[1,1] 2066 1.000
## Gamma[1,2] 1987 1.000
## Gamma[1,3] 4086 1.001
## Gamma[2,1] 1333 1.002
## Gamma[2,2] 1526 1.002
## Gamma[2,3] 3405 1.000
## Gamma[3,1] 3718 1.001
## Gamma[3,2] 4353 1.000
## Gamma[3,3] 3762 1.001
## lambda[1] 1953 1.002
## lambda[2] 1658 1.000
## lambda[3] 2877 1.001
## ta[1,1] 2066 1.000
## ta[1,2] 1987 1.000
## ta[1,3] 4086 1.001
## ta[2,1] 1333 1.002
## ta[2,2] 1526 1.002
## ta[2,3] 3405 1.000
## ta[3,1] 3718 1.001
## ta[3,2] 4353 1.000
## ta[3,3] 3762 1.001
## statdist[1] 5011 1.000
## statdist[2] 3710 1.001
## statdist[3] 4474 1.001
## lp__ 1025 1.004
##
## Samples were drawn using NUTS(diag_e) at Fri Jun 16 13:19:35 2023.
## For each parameter, n_eff is a crude measure of effective sample size,
## and Rhat is the potential scale reduction factor on split chains (at
## convergence, Rhat=1).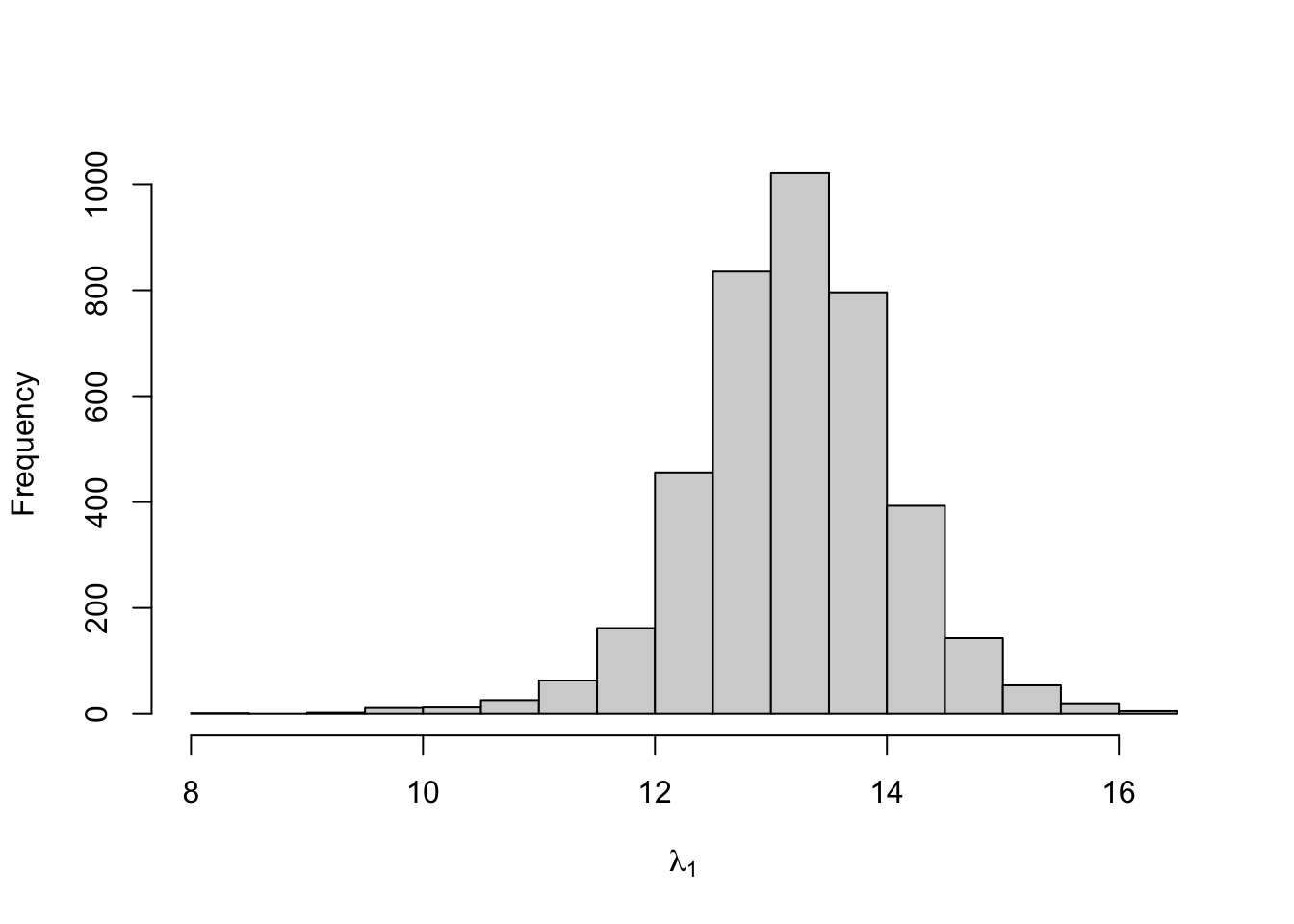
Figure 8.13: Posterior distribution for the state-dependent means of the three-state Poisson-HMM.
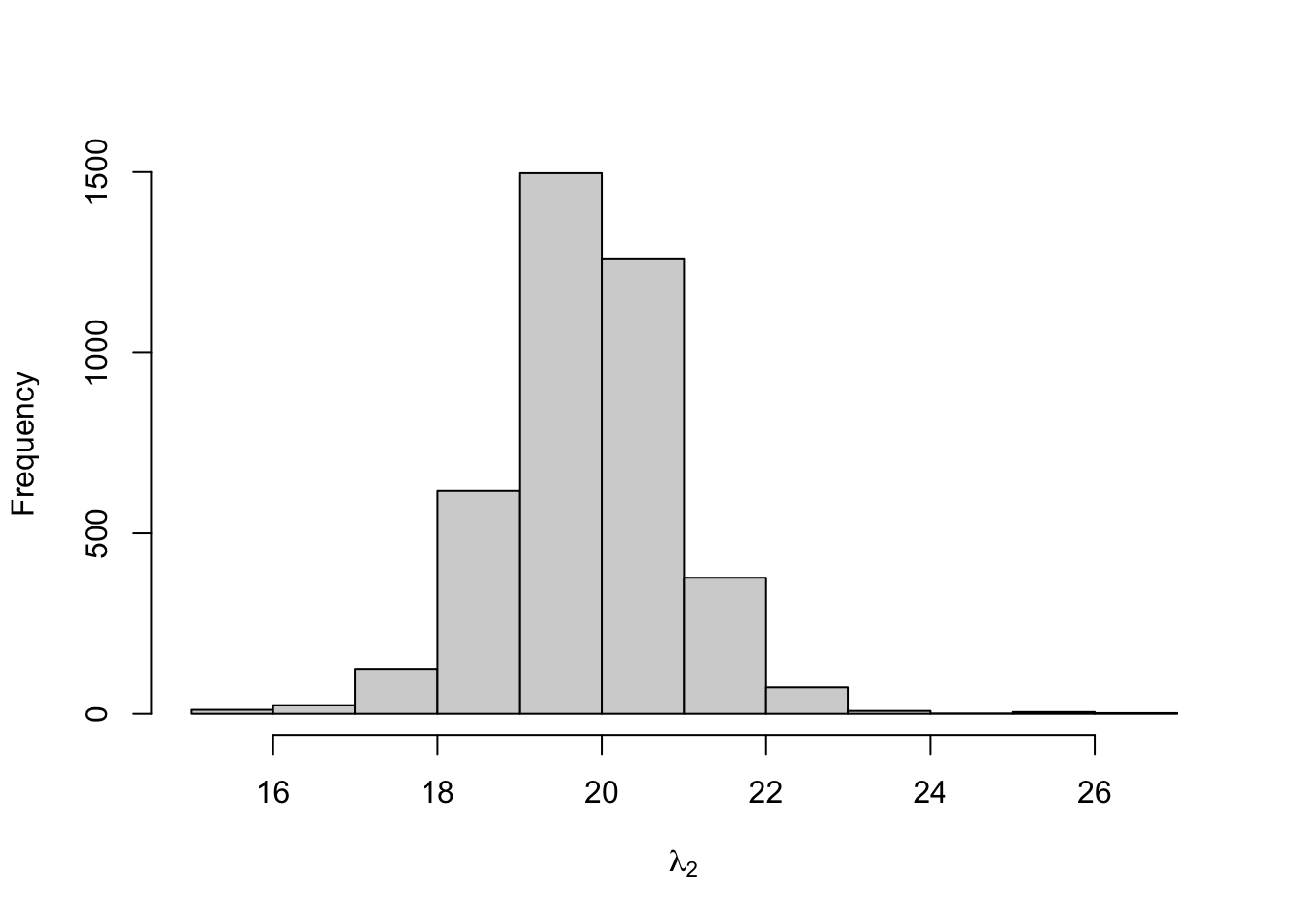
Figure 8.14: Posterior distribution for the state-dependent means of the three-state Poisson-HMM.
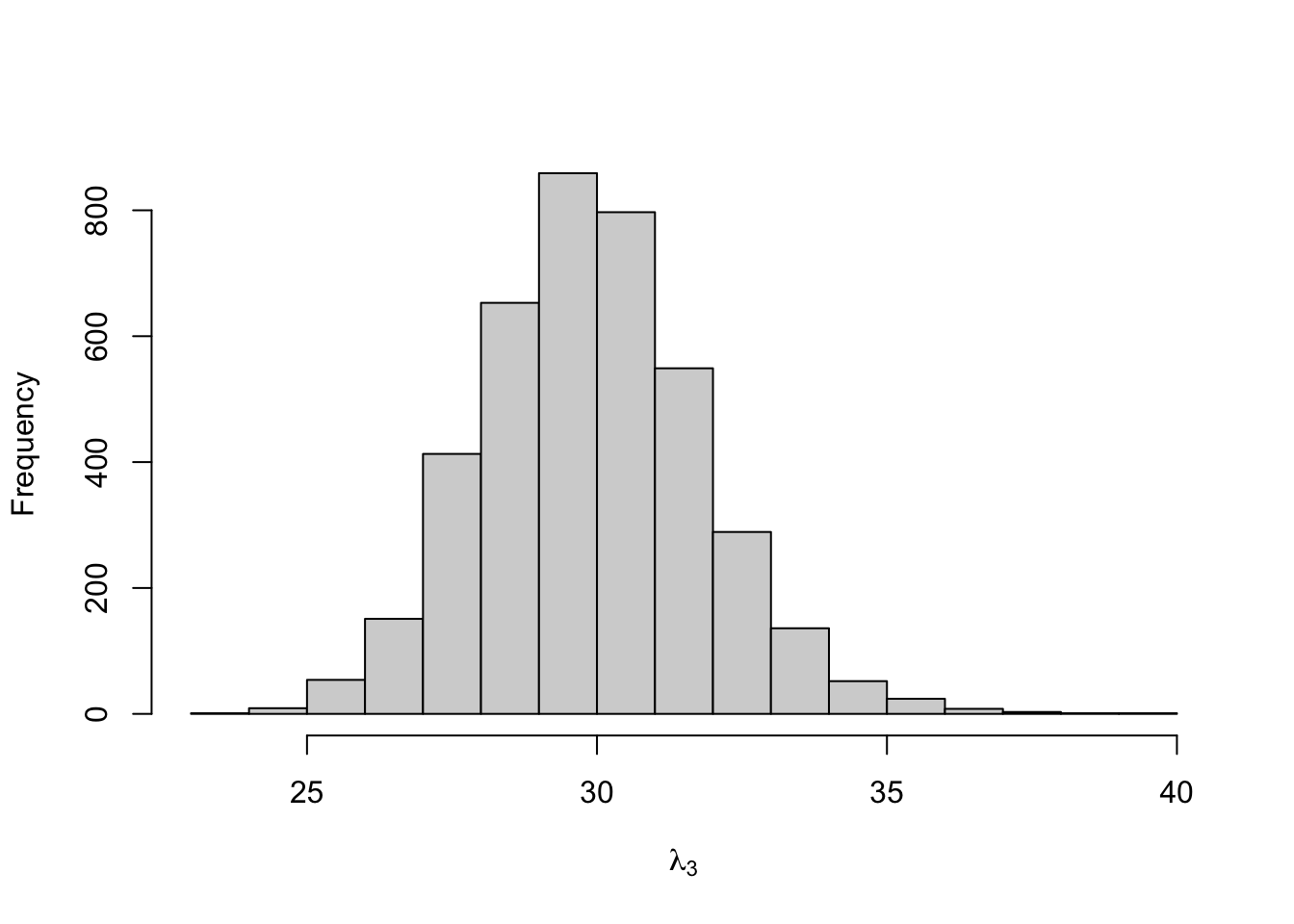
Figure 8.15: Posterior distribution for the state-dependent means of the three-state Poisson-HMM.
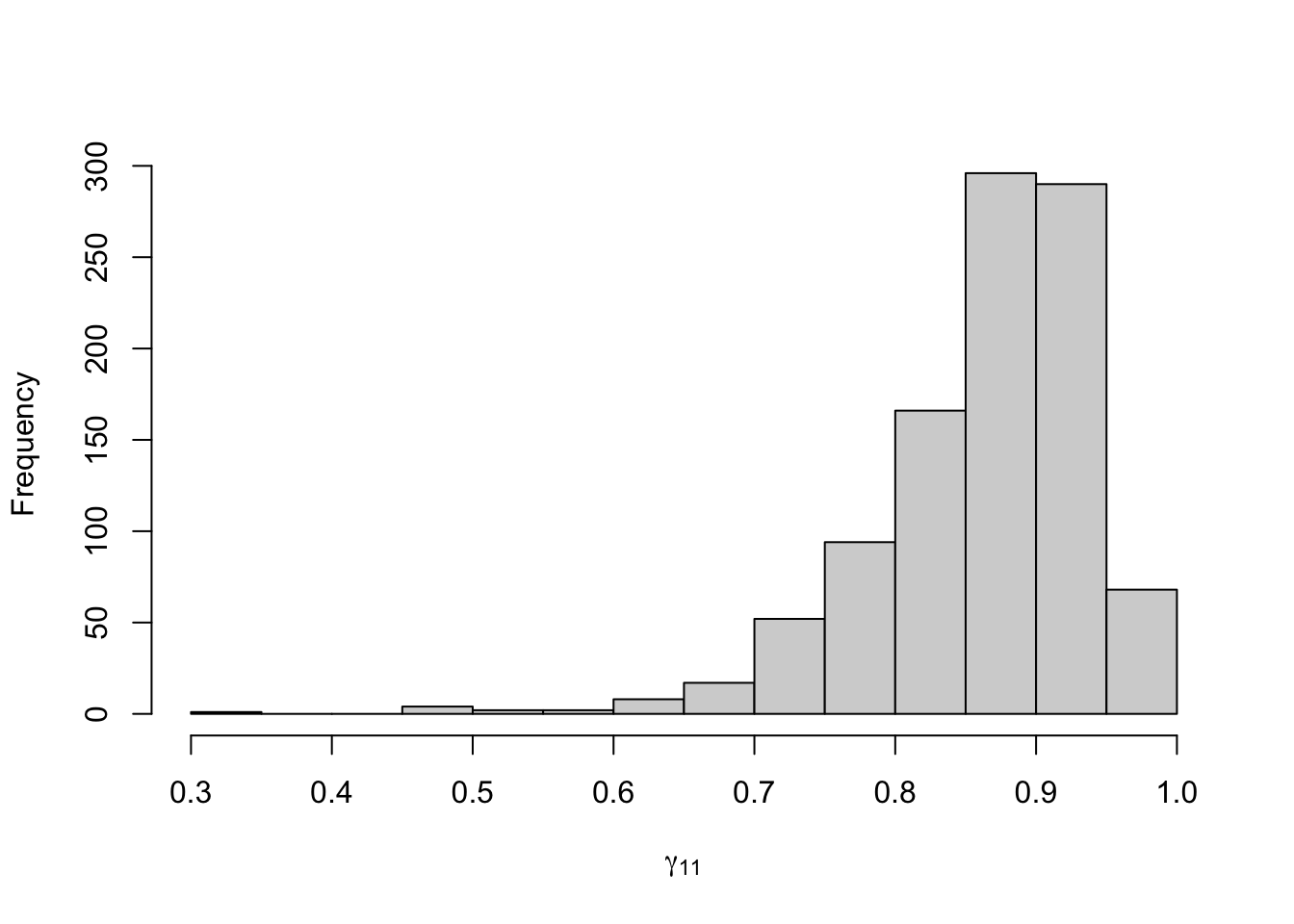
Figure 8.16: Posterior distribution for the transition probabilities of the three-state Poisson-HMM.
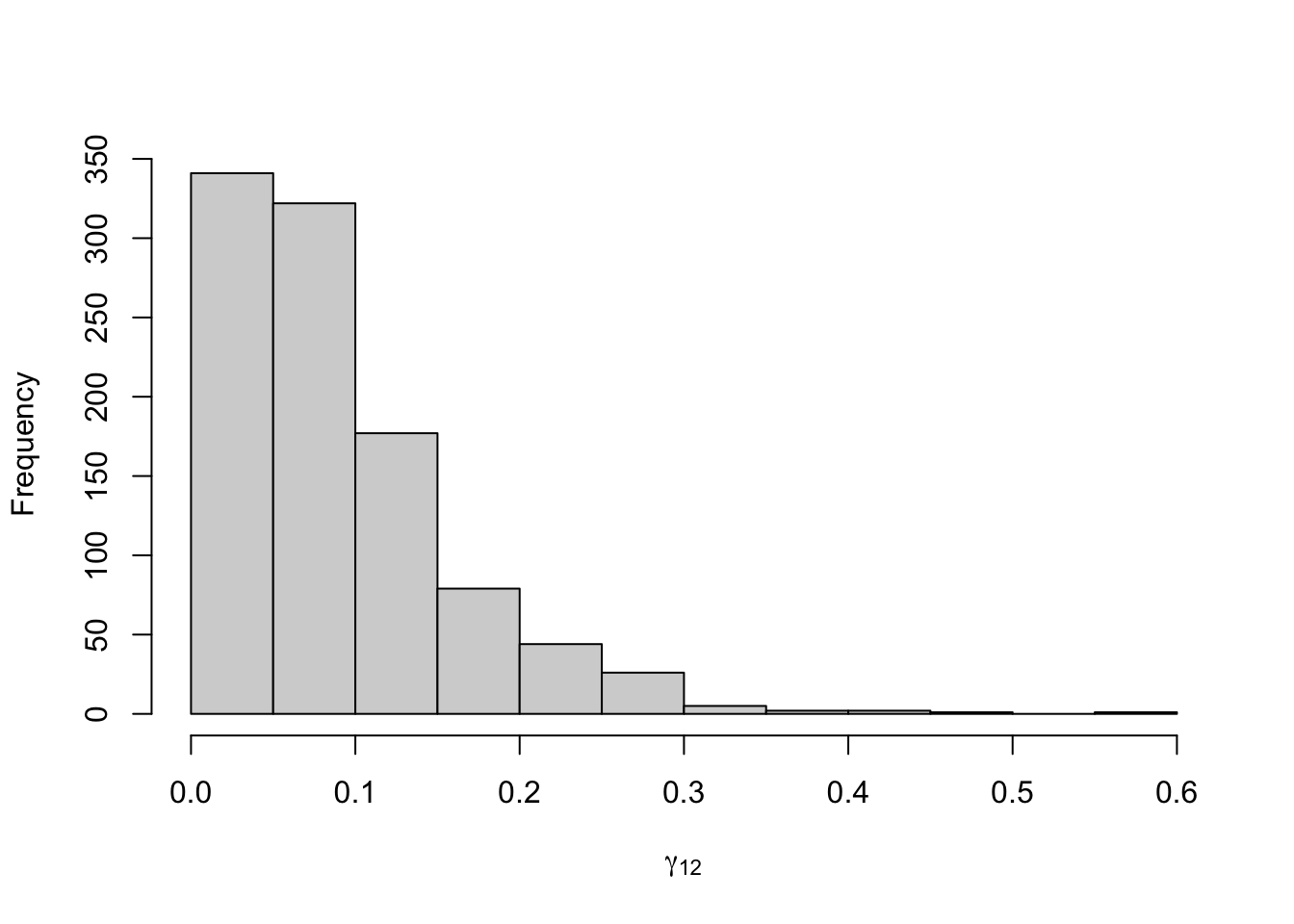
Figure 8.17: Posterior distribution for the transition probabilities of the three-state Poisson-HMM.
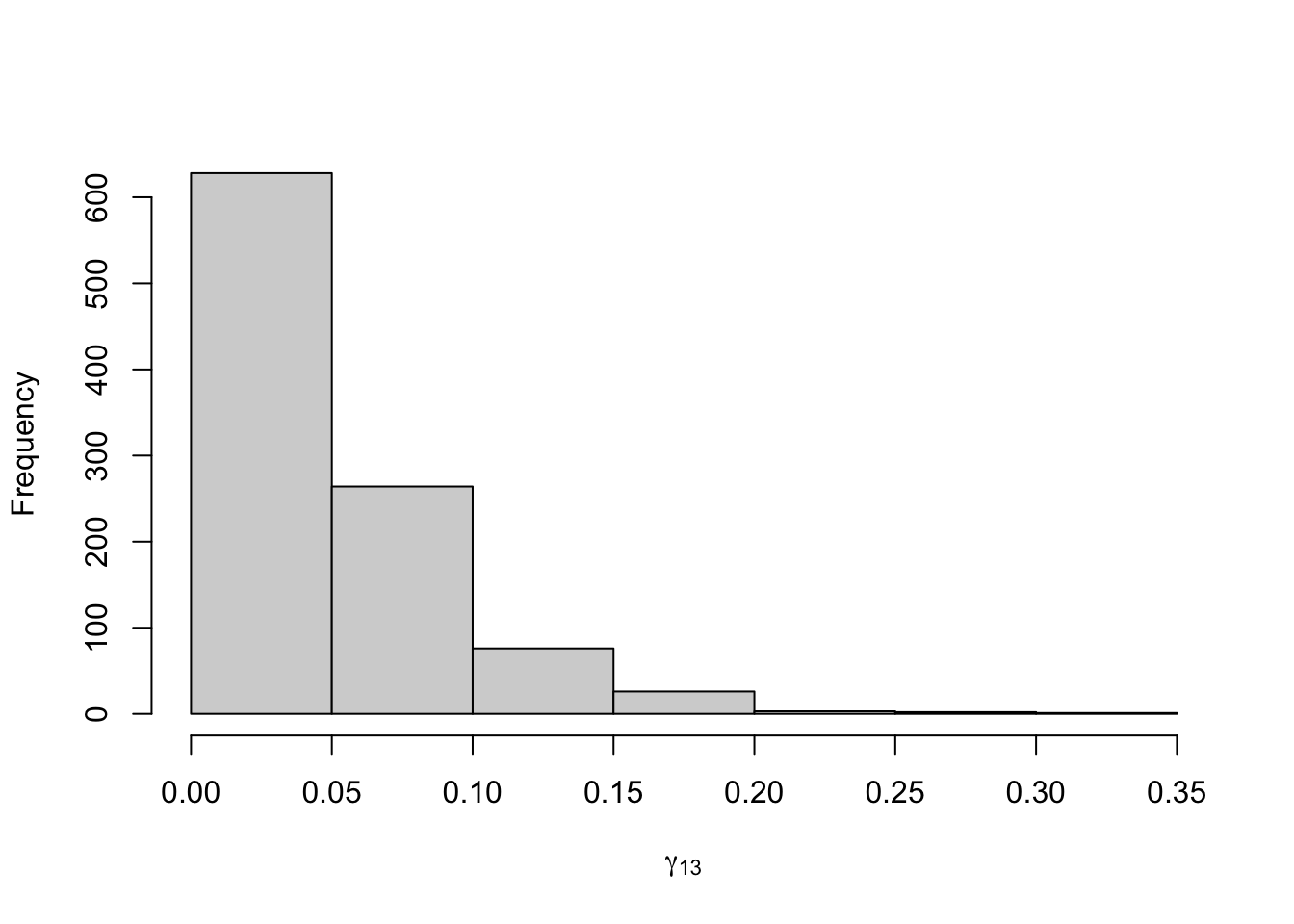
Figure 8.18: Posterior distribution for the transition probabilities of the three-state Poisson-HMM.
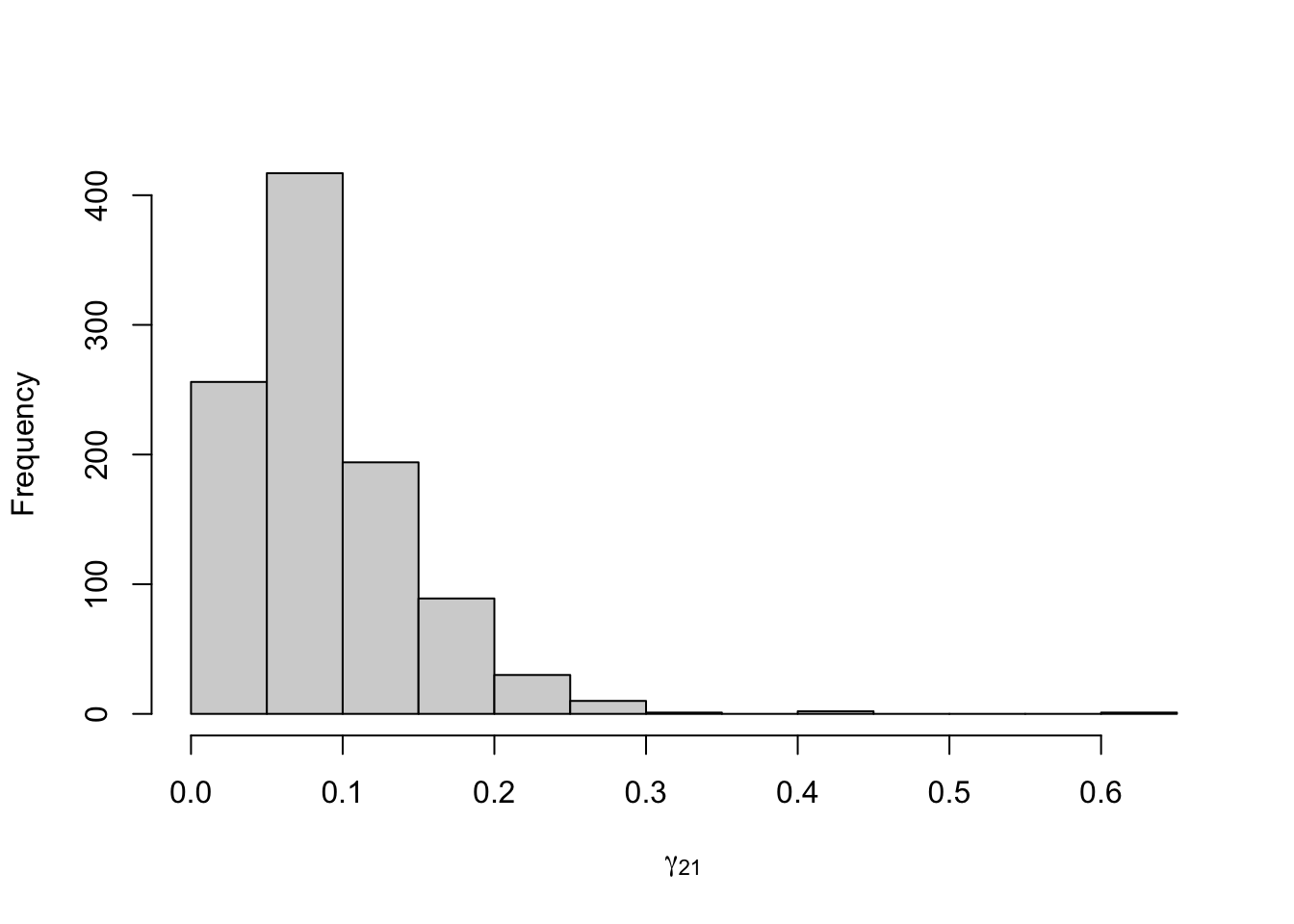
Figure 8.19: Posterior distribution for the transition probabilities of the three-state Poisson-HMM.
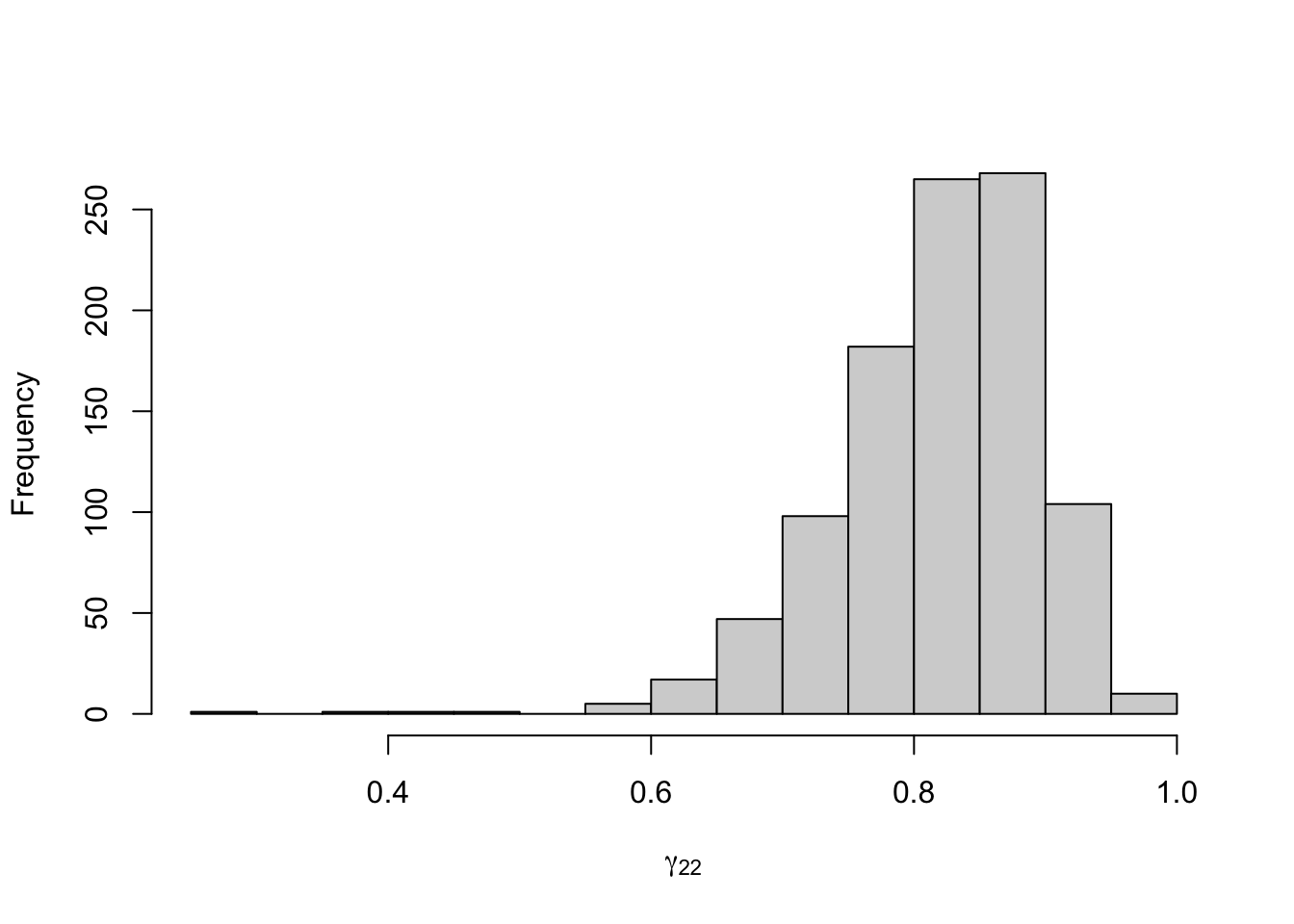
Figure 8.20: Posterior distribution for the transition probabilities of the three-state Poisson-HMM.
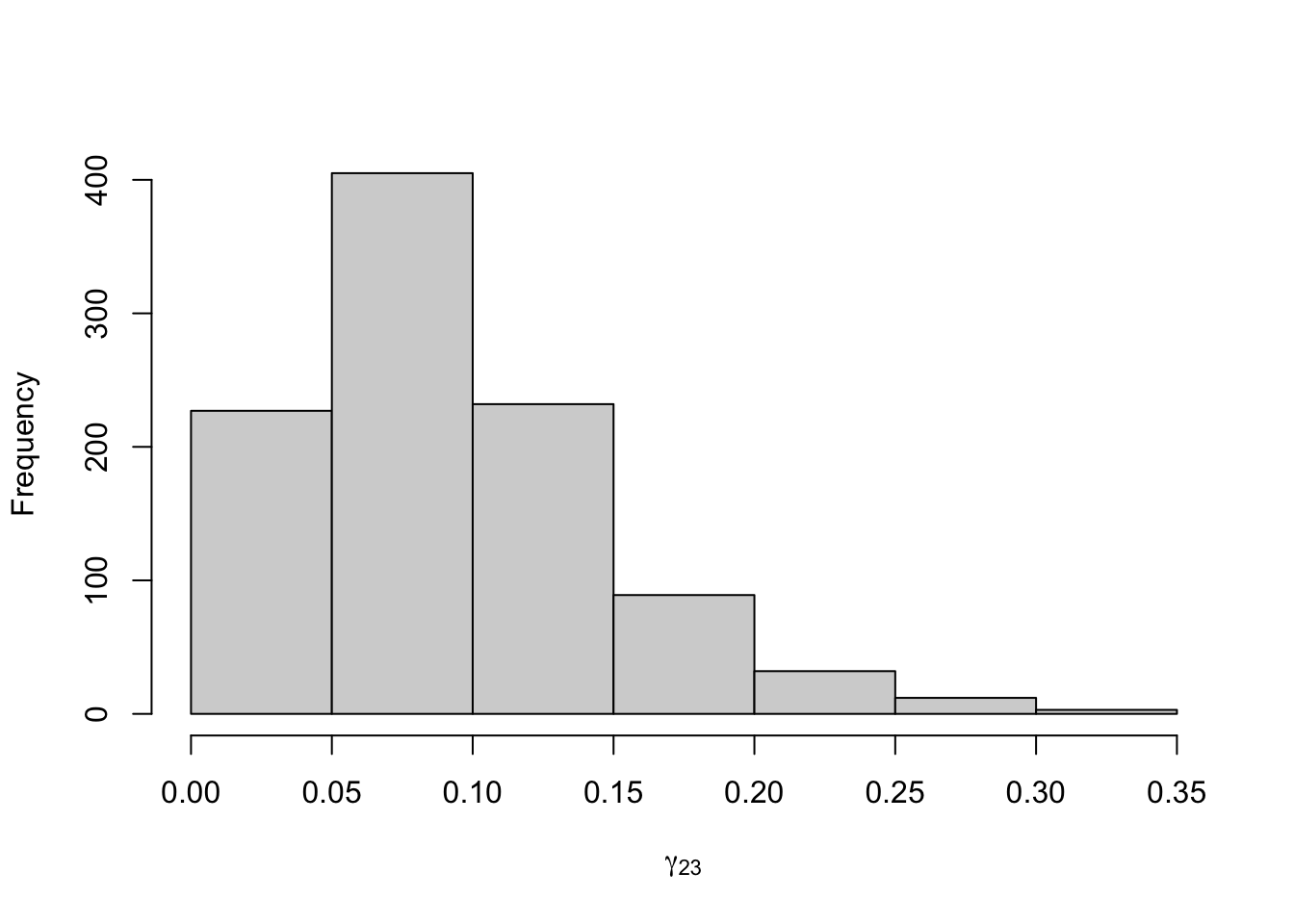
Figure 8.21: Posterior distribution for the transition probabilities of the three-state Poisson-HMM.
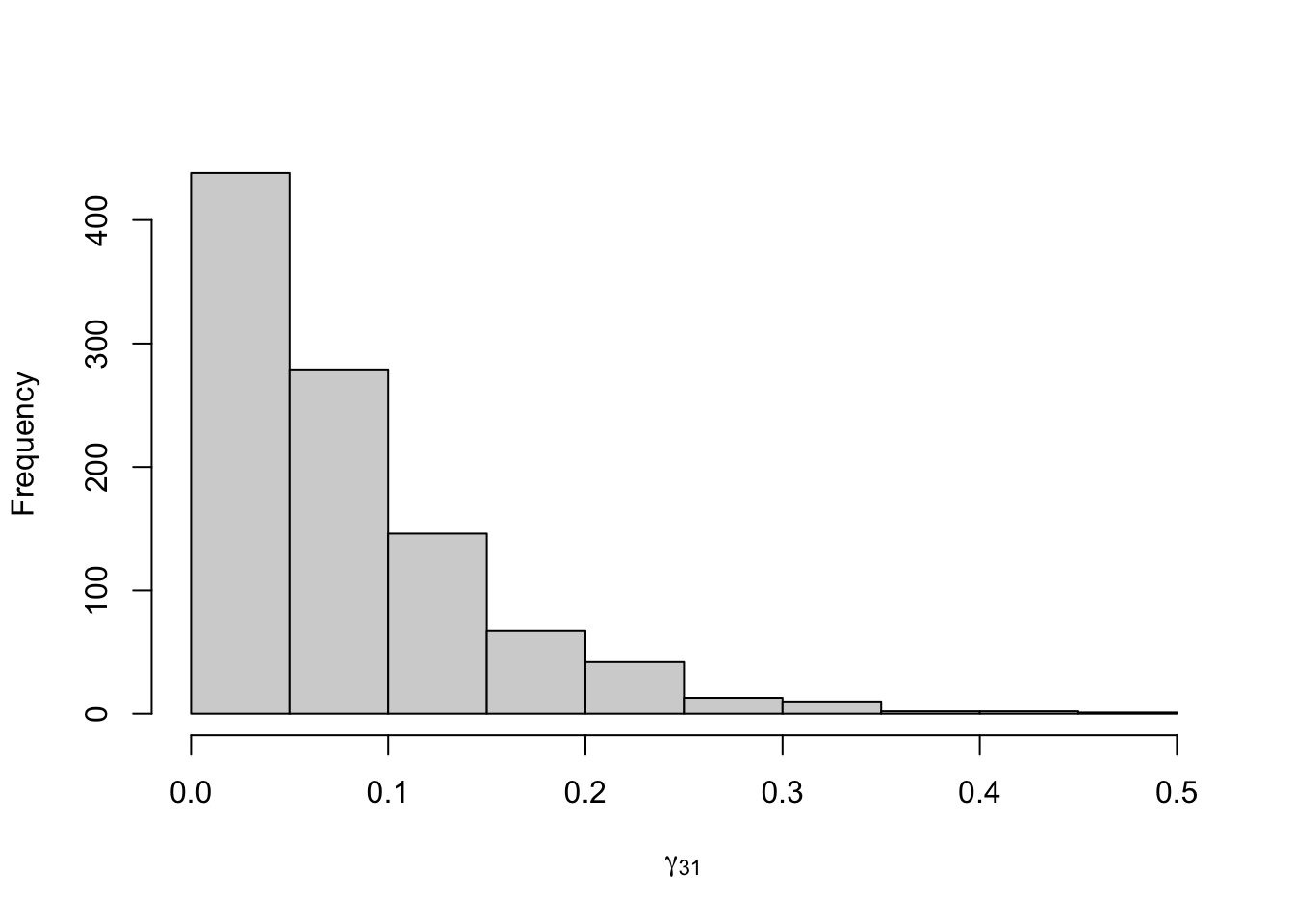
Figure 8.22: Posterior distribution for the transition probabilities of the three-state Poisson-HMM.
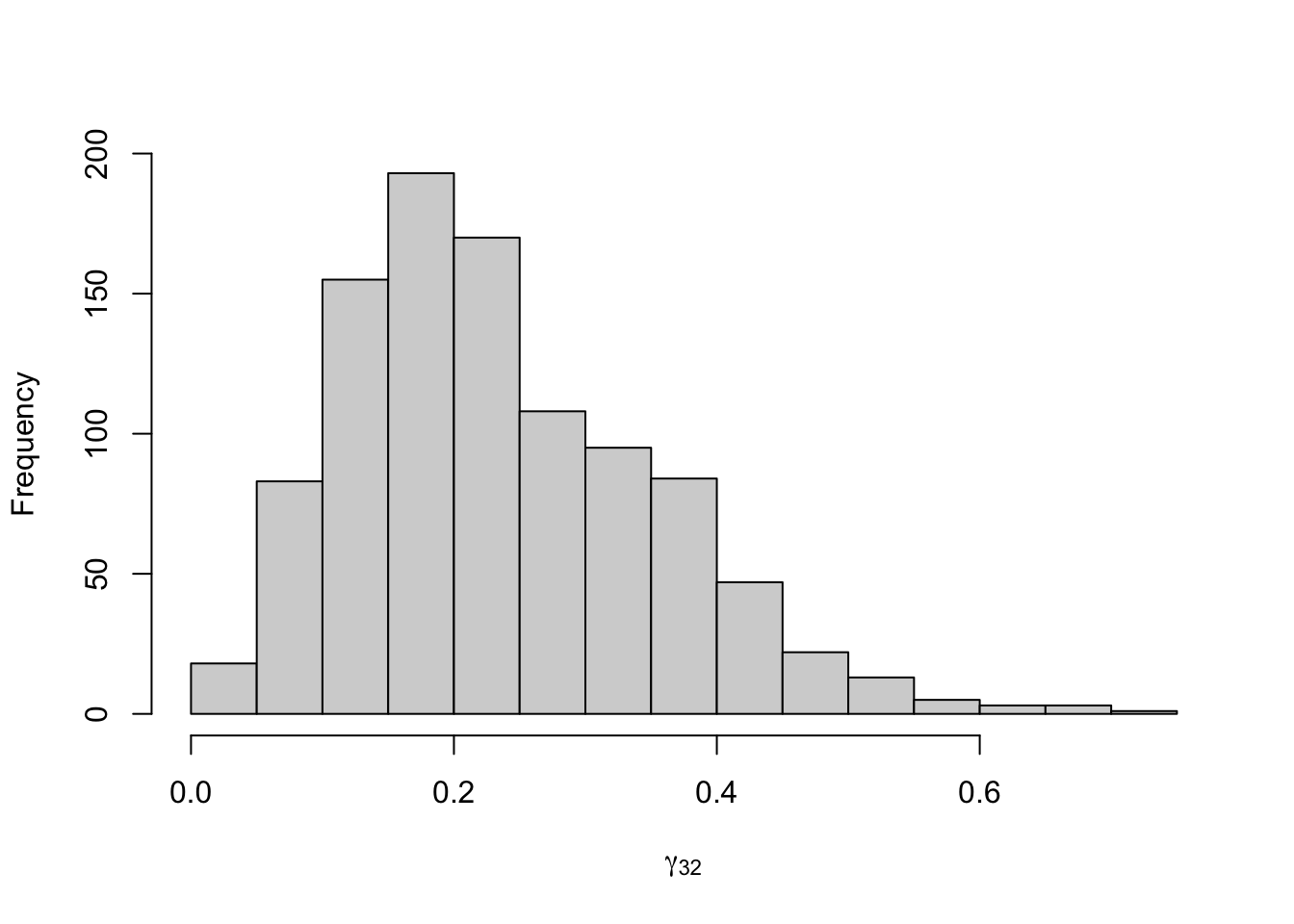
Figure 8.23: Posterior distribution for the transition probabilities of the three-state Poisson-HMM.
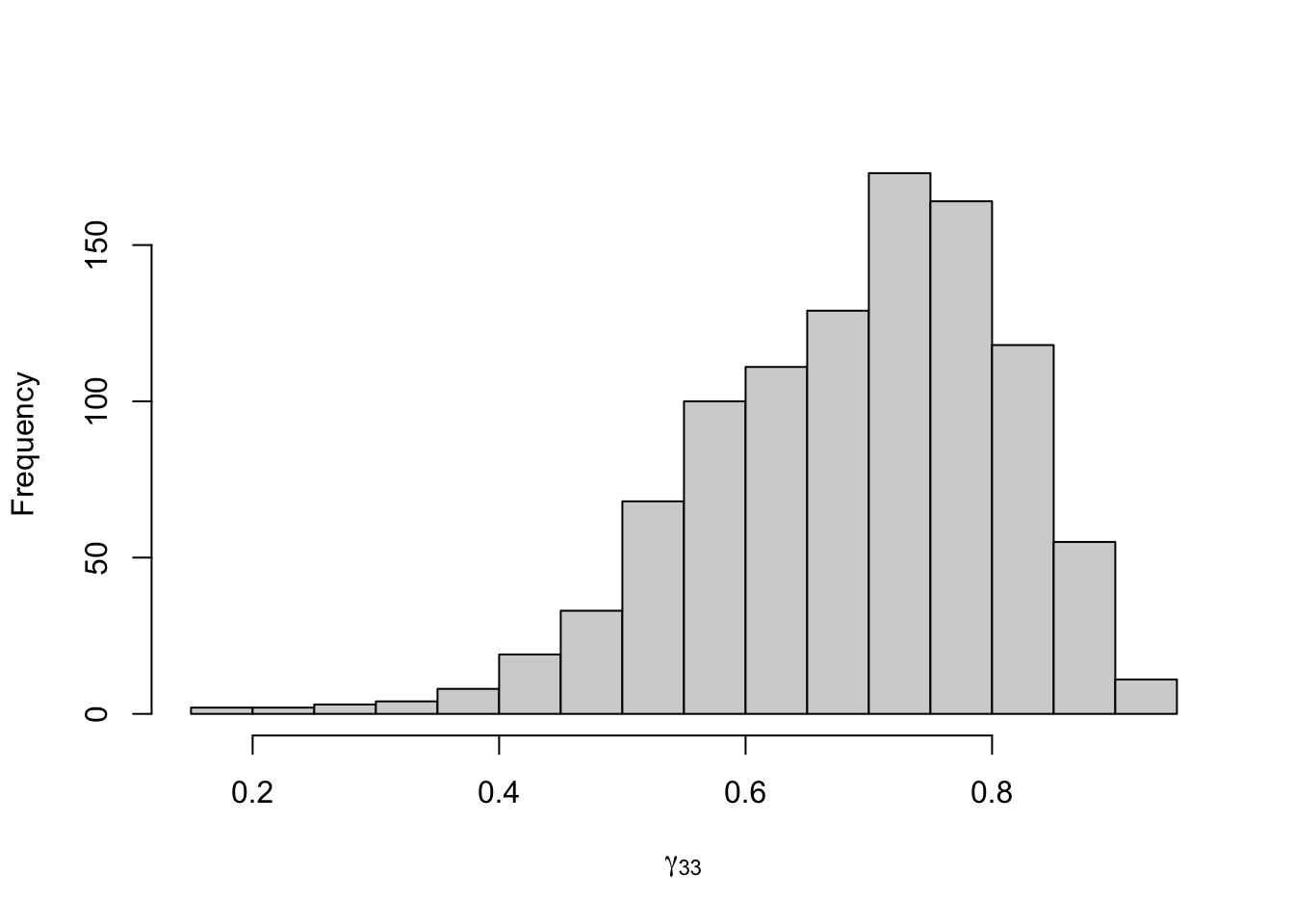
Figure 8.24: Posterior distribution for the transition probabilities of the three-state Poisson-HMM.
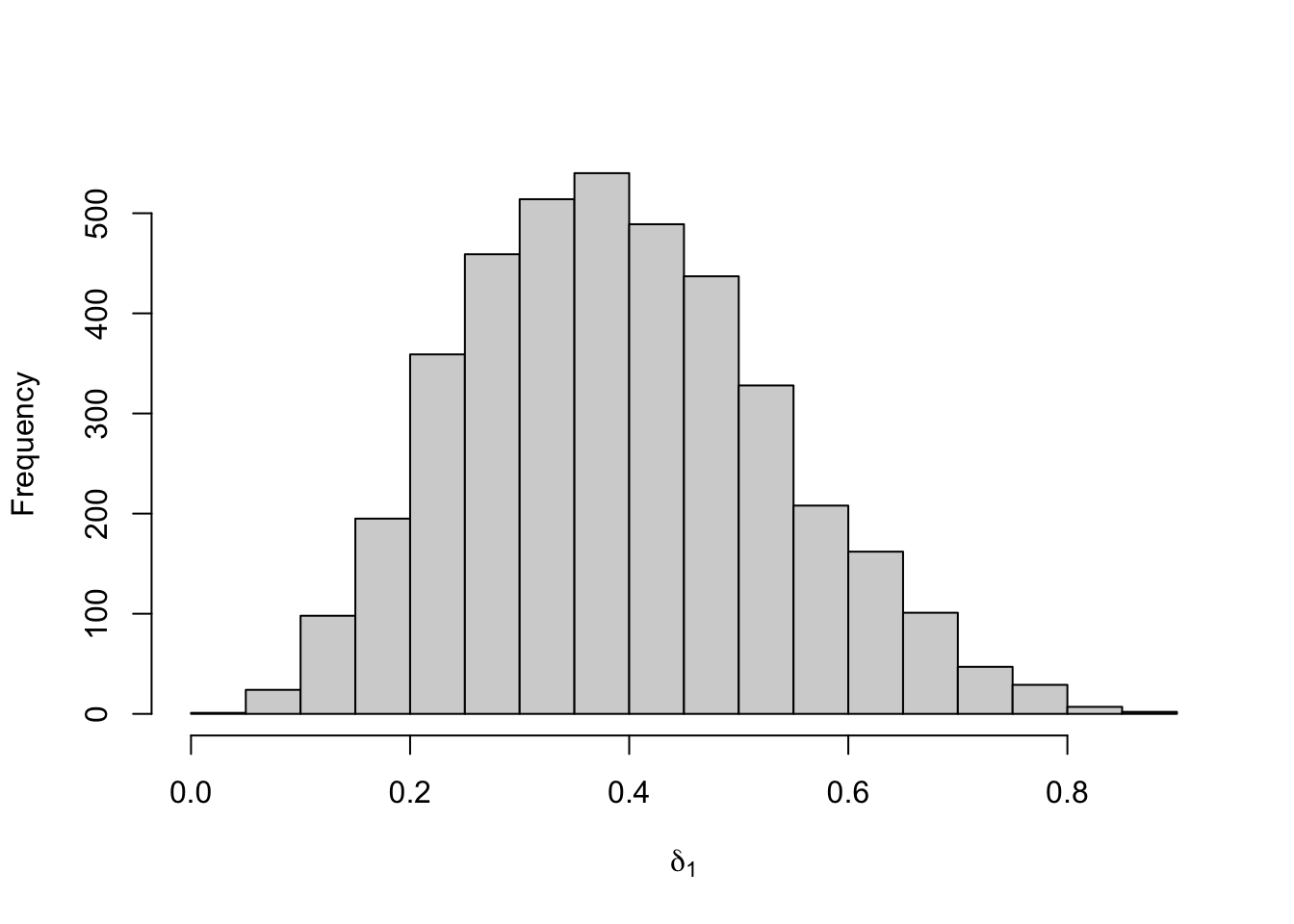
Figure 8.25: Posterior distribution for the initial distribution of the three-state Poisson-HMM.
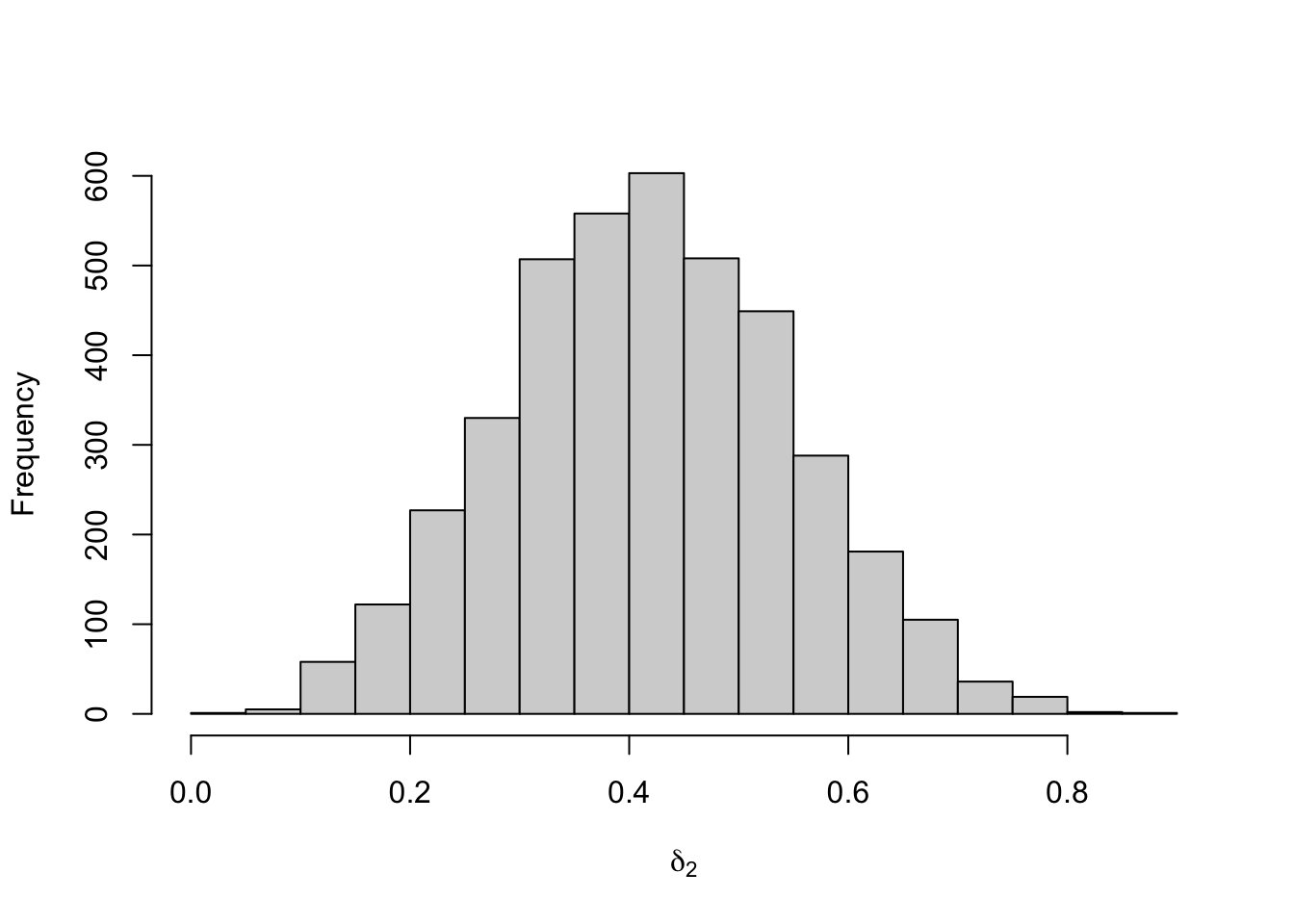
Figure 8.26: Posterior distribution for the initial distribution of the three-state Poisson-HMM.
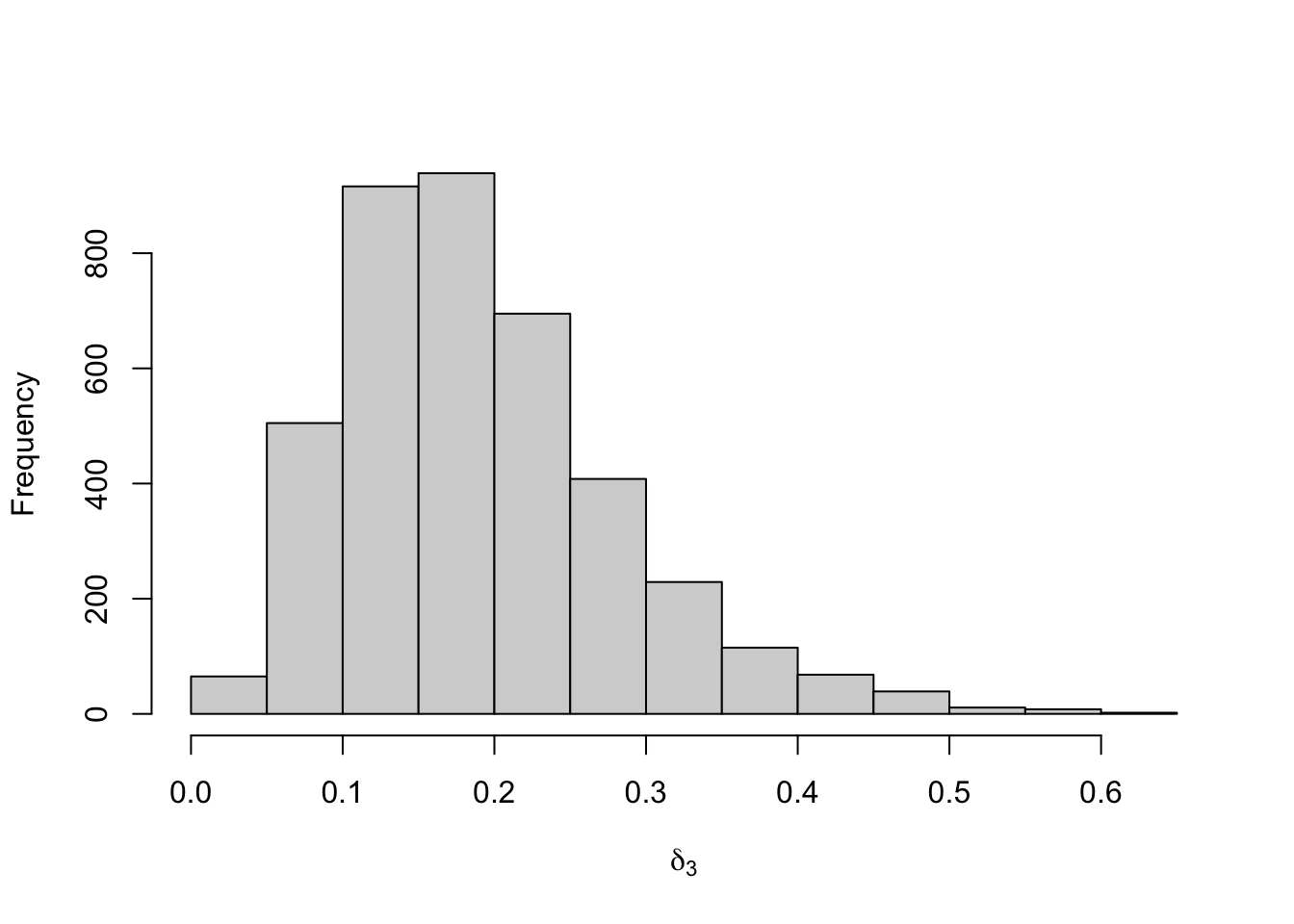
Figure 8.27: Posterior distribution for the initial distribution of the three-state Poisson-HMM.